Deposition Date
2015-11-23
Release Date
2015-12-30
Last Version Date
2023-09-27
Entry Detail
PDB ID:
5EXD
Keywords:
Title:
Crystal structure of oxalate oxidoreductase from Moorella thermoacetica bound with carboxy-di-oxido-methyl-TPP (COOM-TPP) intermediate
Biological Source:
Source Organism(s):
Moorella thermoacetica (strain ATCC 39073) (Taxon ID: 264732)
Method Details:
Experimental Method:
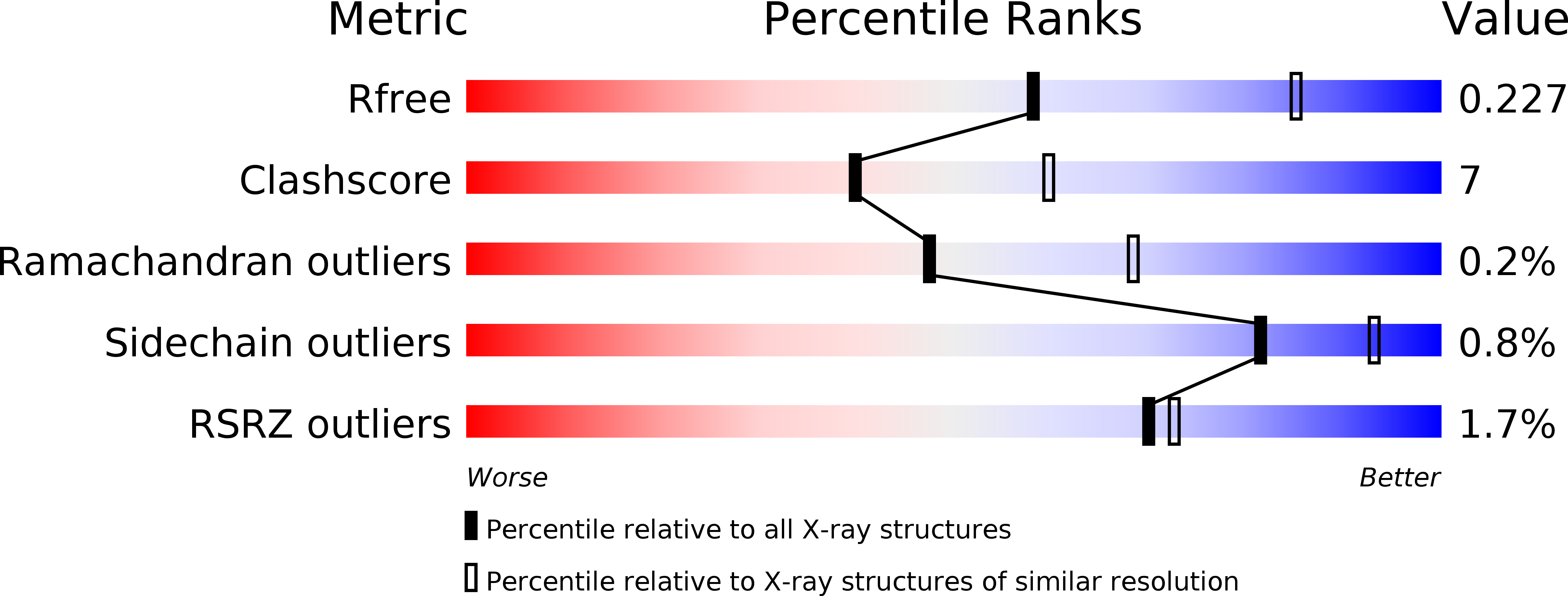
Resolution:
2.50 Å
R-Value Free:
0.22
R-Value Work:
0.19
R-Value Observed:
0.20
Space Group:
P 43