Deposition Date
2015-10-09
Release Date
2016-05-18
Last Version Date
2025-04-02
Entry Detail
PDB ID:
5E5T
Keywords:
Title:
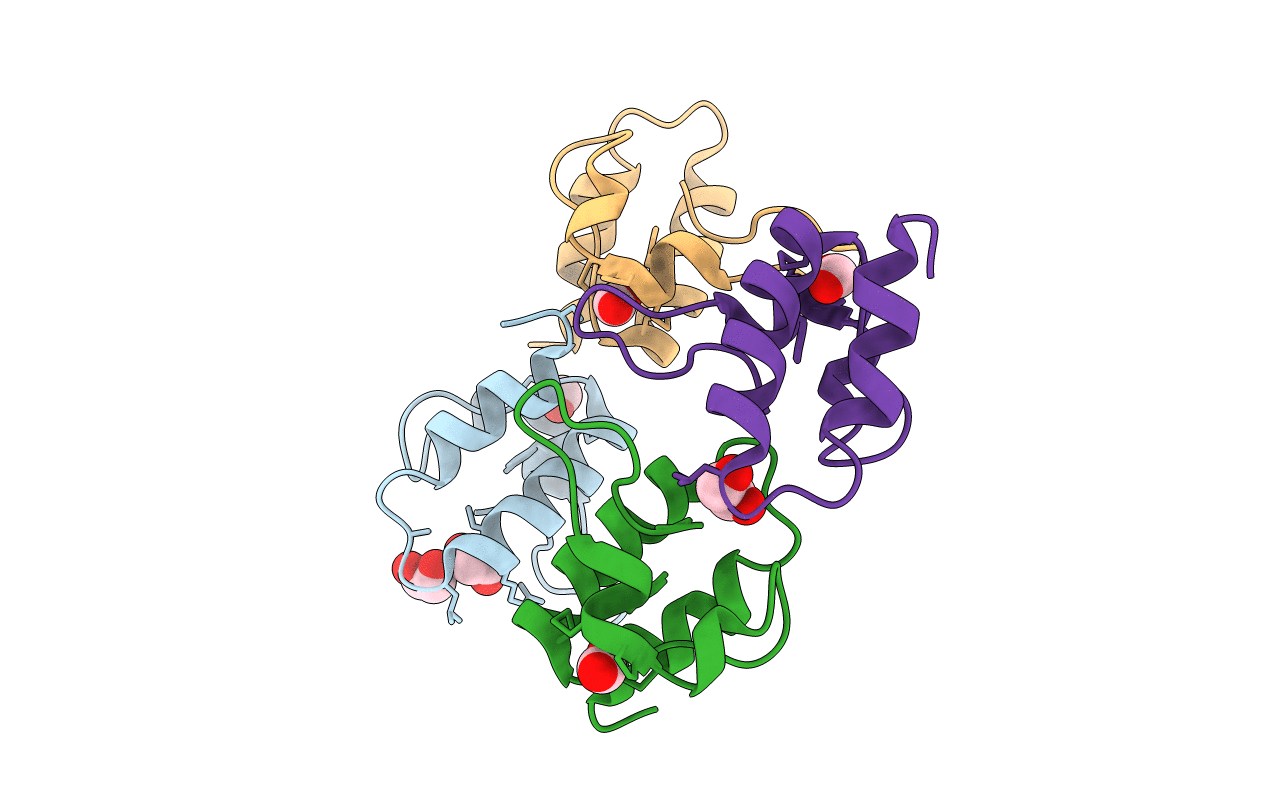
Quasi-racemic snakin-1 in P1 after radiation damage
Biological Source:
Source Organism(s):
Solanum tuberosum (Taxon ID: 4113)
Method Details:
Experimental Method:
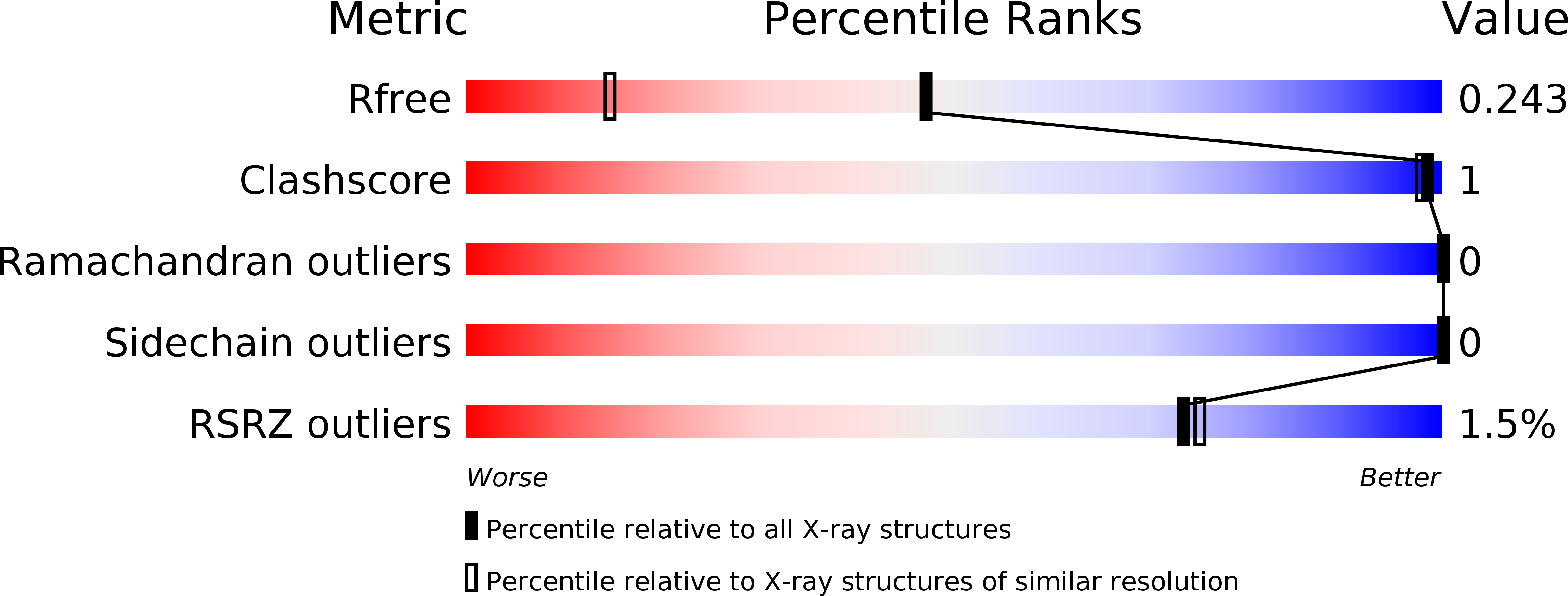
Resolution:
1.57 Å
R-Value Free:
0.24
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
P 1