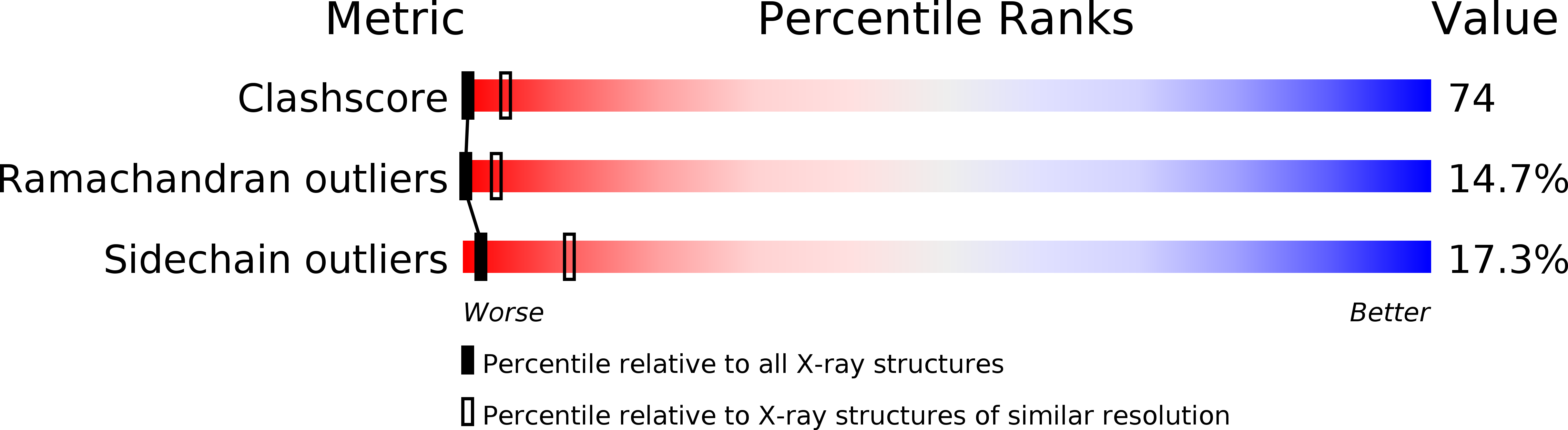
Deposition Date
2015-08-25
Release Date
2015-09-23
Last Version Date
2024-01-10
Entry Detail
PDB ID:
5DDJ
Keywords:
Title:
Crystal structure of recombinant foot-and-mouth-disease virus O1M-S2093Y empty capsid
Biological Source:
Source Organism(s):
Foot-and-mouth disease virus - type O (Taxon ID: 12118)
Expression System(s):
Method Details: