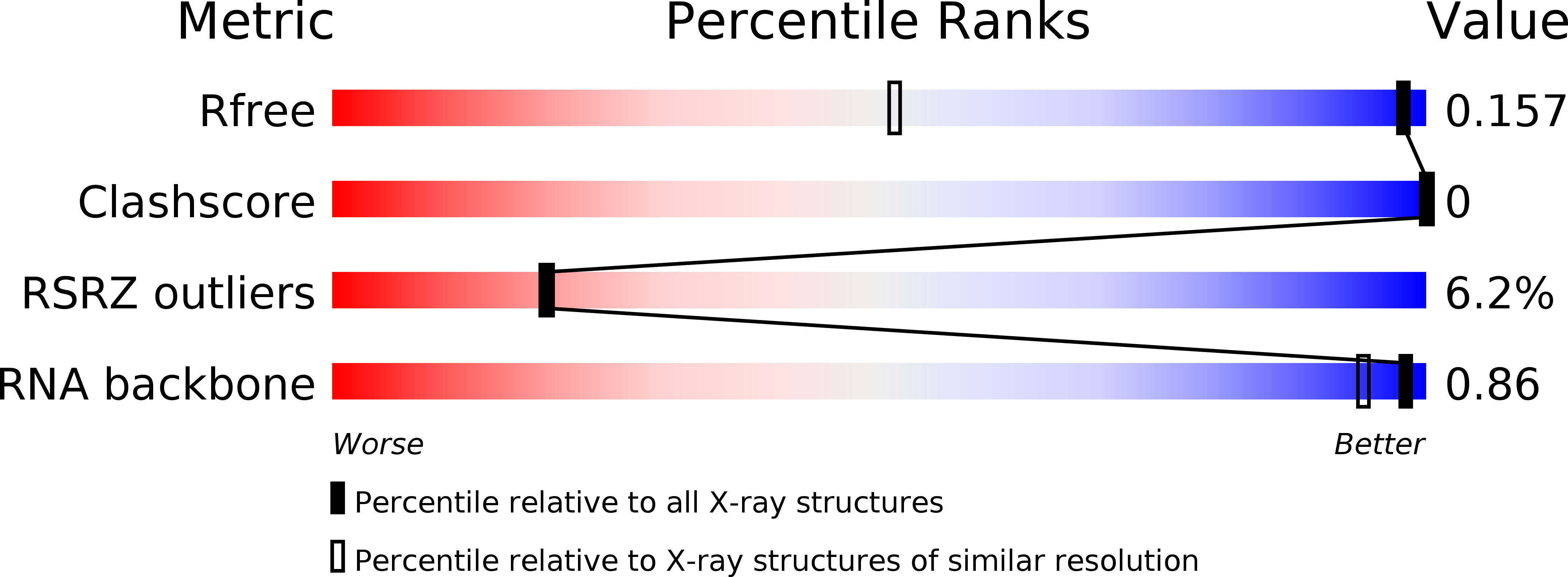Deposition Date
2015-08-19
Release Date
2016-04-13
Last Version Date
2024-03-06
Entry Detail
PDB ID:
5DA6
Keywords:
Title:
Atomic resolution crystal structure of double-stranded RNA 32 base pairs long determined from random starting phases angles in the presence of pseudo translational symmetry using the direct methods program SIR2014.
Biological Source:
Source Organism(s):
Trypanosoma brucei (Taxon ID: 5691)
Method Details:
Experimental Method:
Resolution:
1.05 Å
R-Value Free:
0.15
R-Value Work:
0.12
R-Value Observed:
0.13
Space Group:
H 3 2