Deposition Date
2015-07-15
Release Date
2016-03-02
Last Version Date
2023-09-27
Entry Detail
PDB ID:
5CKR
Keywords:
Title:
Crystal Structure of MraY in complex with Muraymycin D2
Biological Source:
Source Organism:
Aquifex aeolicus (strain VF5) (Taxon ID: 224324)
Host Organism:
Method Details:
Experimental Method:
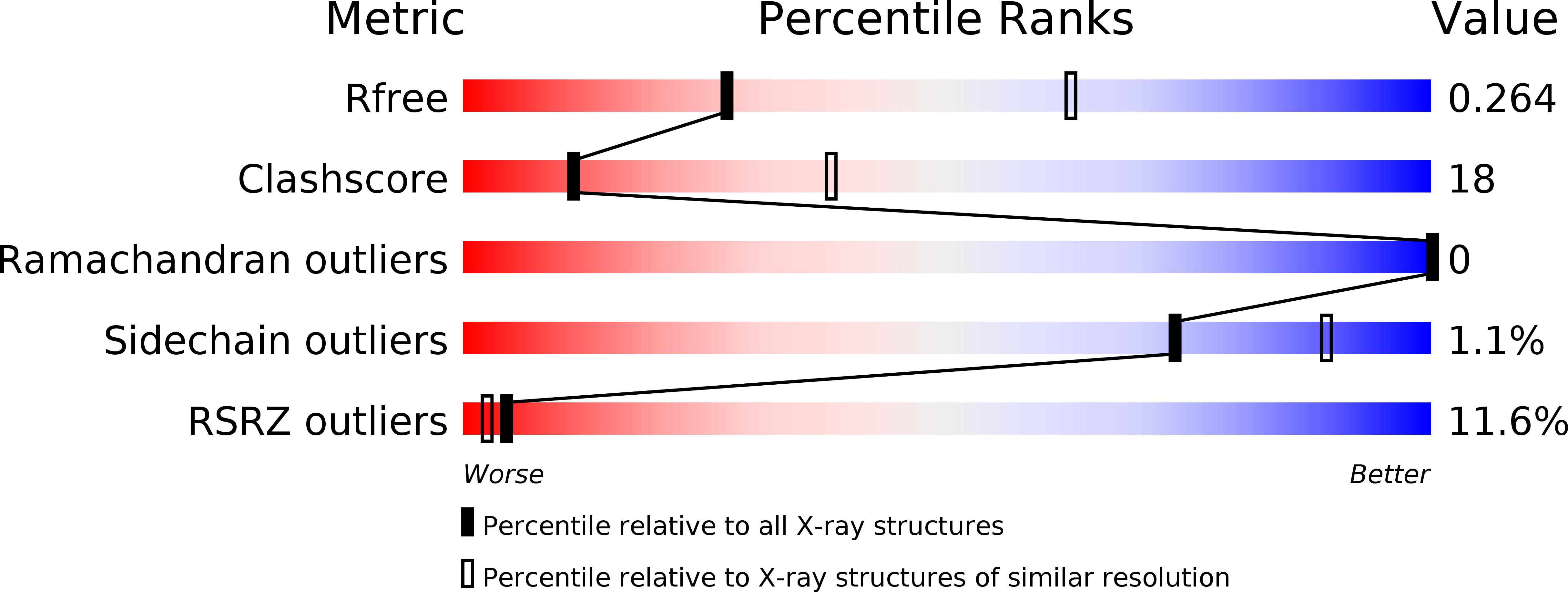
Resolution:
2.95 Å
R-Value Free:
0.26
R-Value Work:
0.24
R-Value Observed:
0.24
Space Group:
C 2 2 21