Deposition Date
2015-03-05
Release Date
2015-03-18
Last Version Date
2024-01-10
Entry Detail
PDB ID:
5AKQ
Keywords:
Title:
X-ray structure and mutagenesis studies of the N-isopropylammelide isopropylaminohydrolase, AtzC
Biological Source:
Source Organism:
PSEUDOMONAS SP. ADP (Taxon ID: 47660)
Host Organism:
Method Details:
Experimental Method:
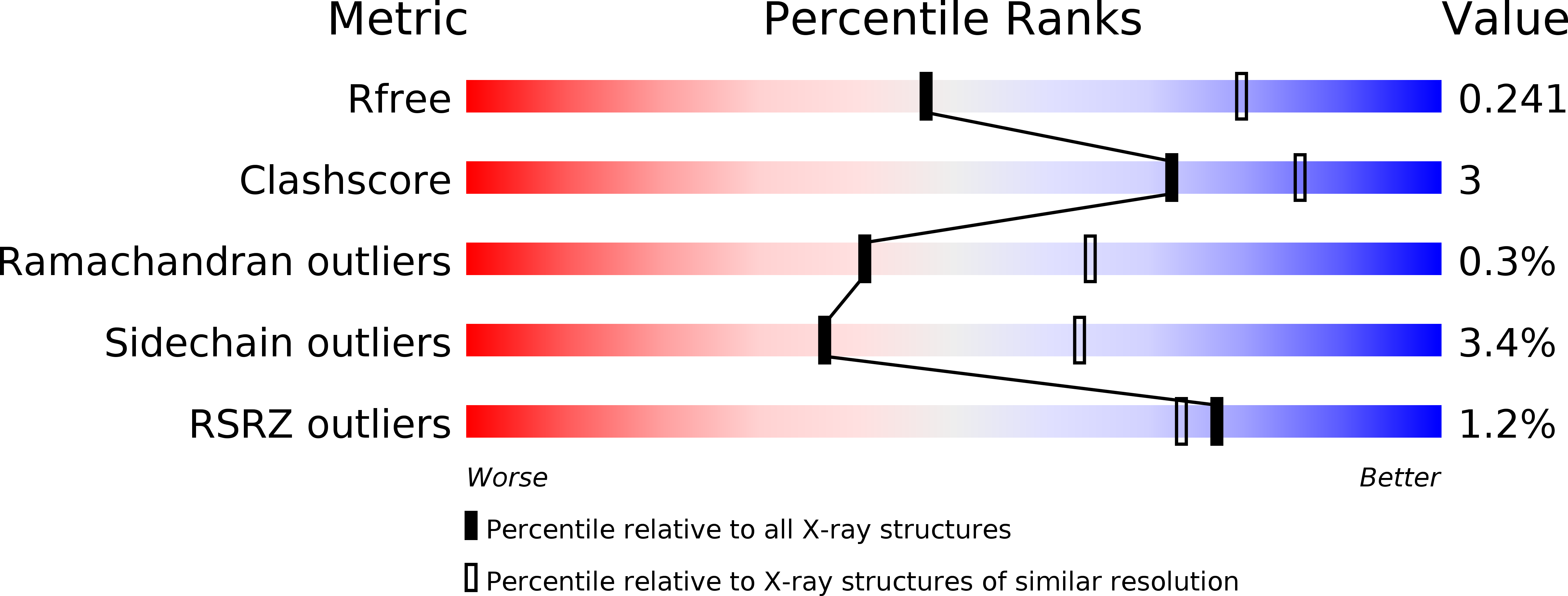
Resolution:
2.60 Å
R-Value Free:
0.24
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
C 1 2 1