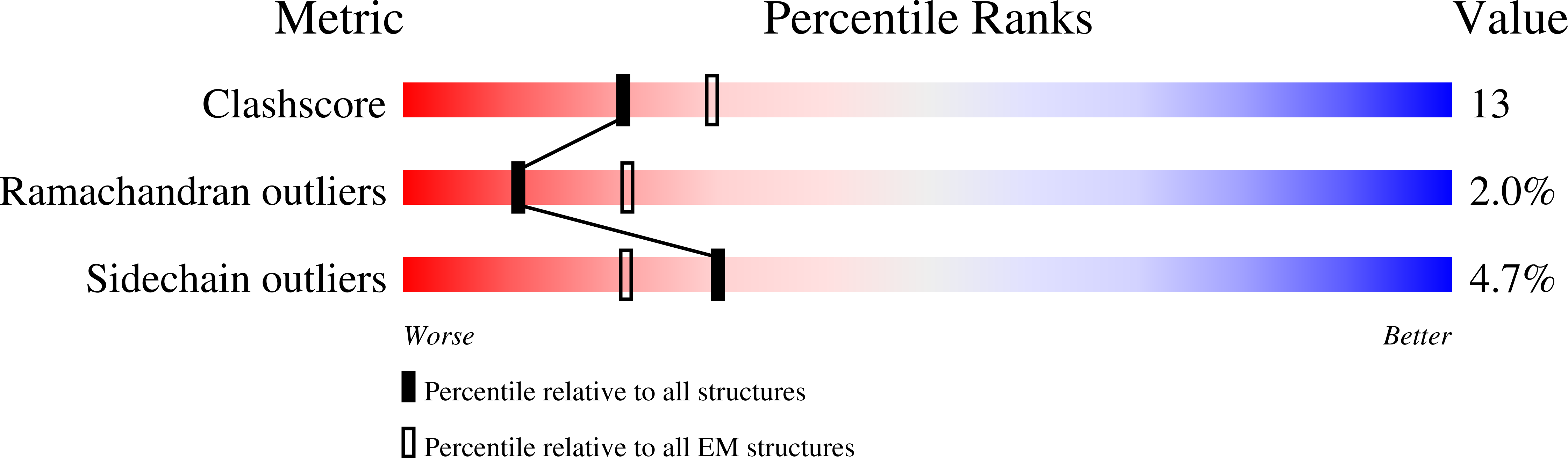
Deposition Date
2015-05-06
Release Date
2015-08-19
Last Version Date
2024-10-23
Entry Detail
PDB ID:
5A22
Keywords:
Title:
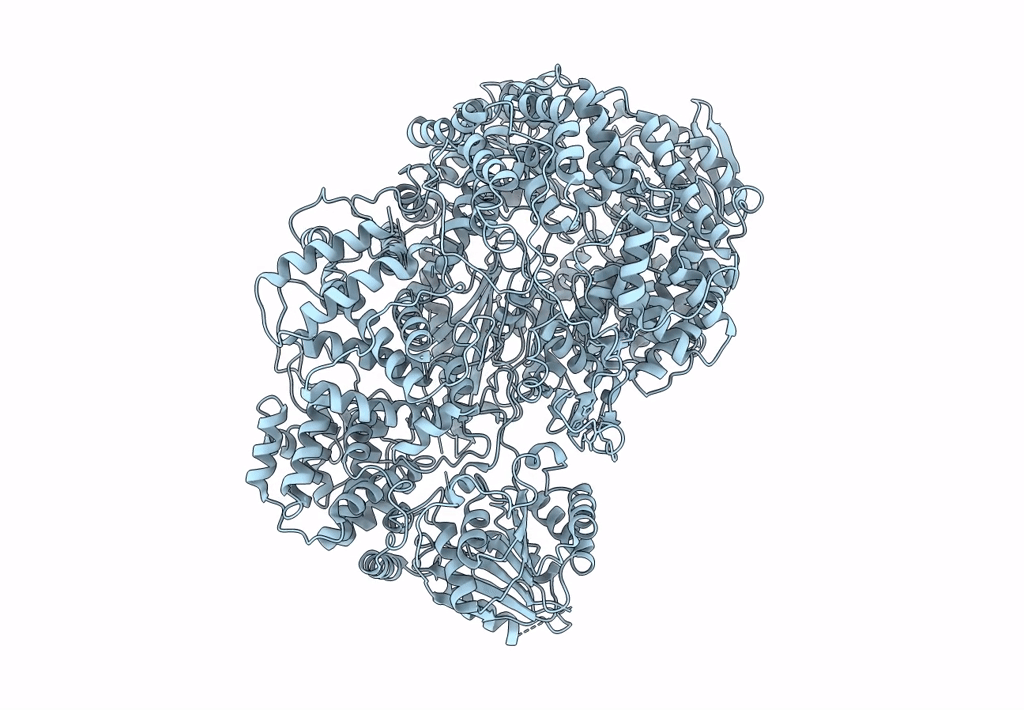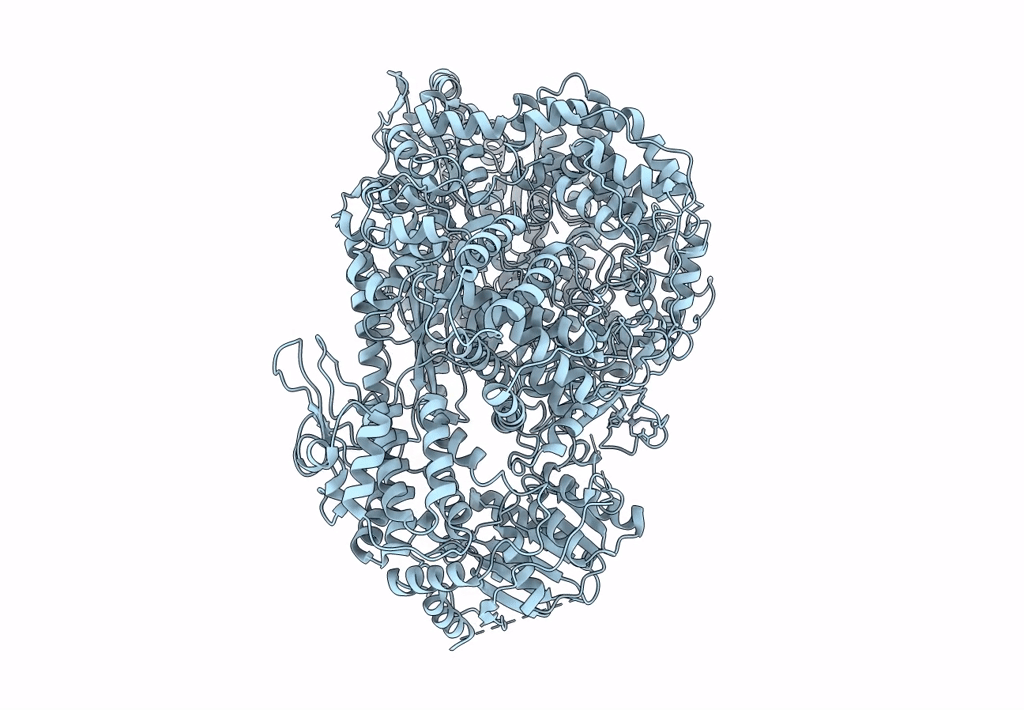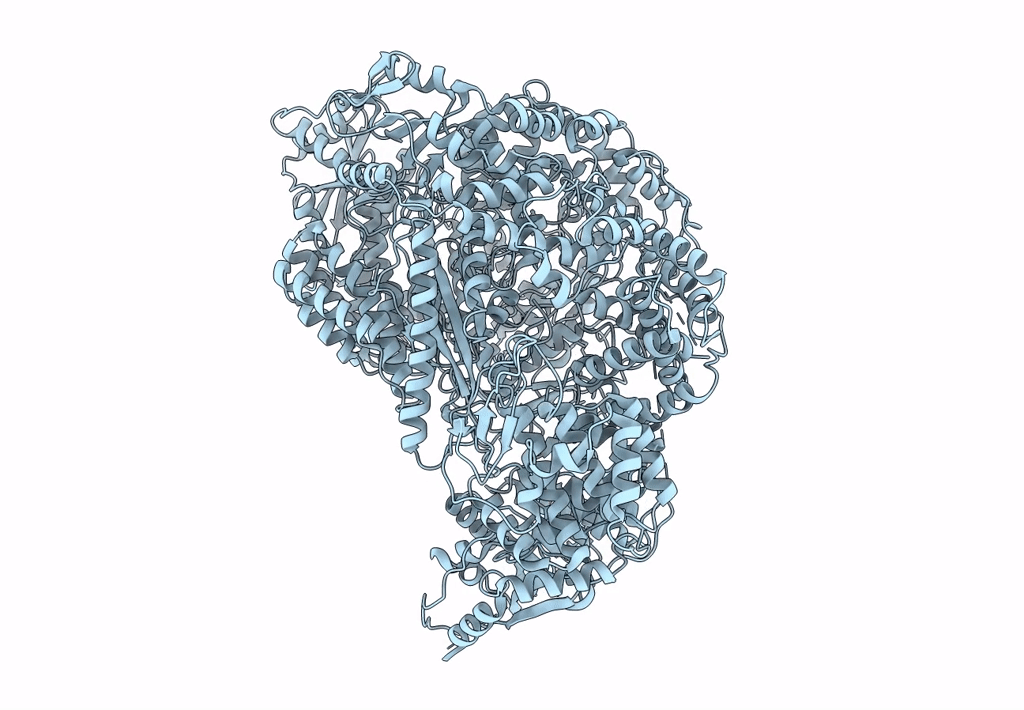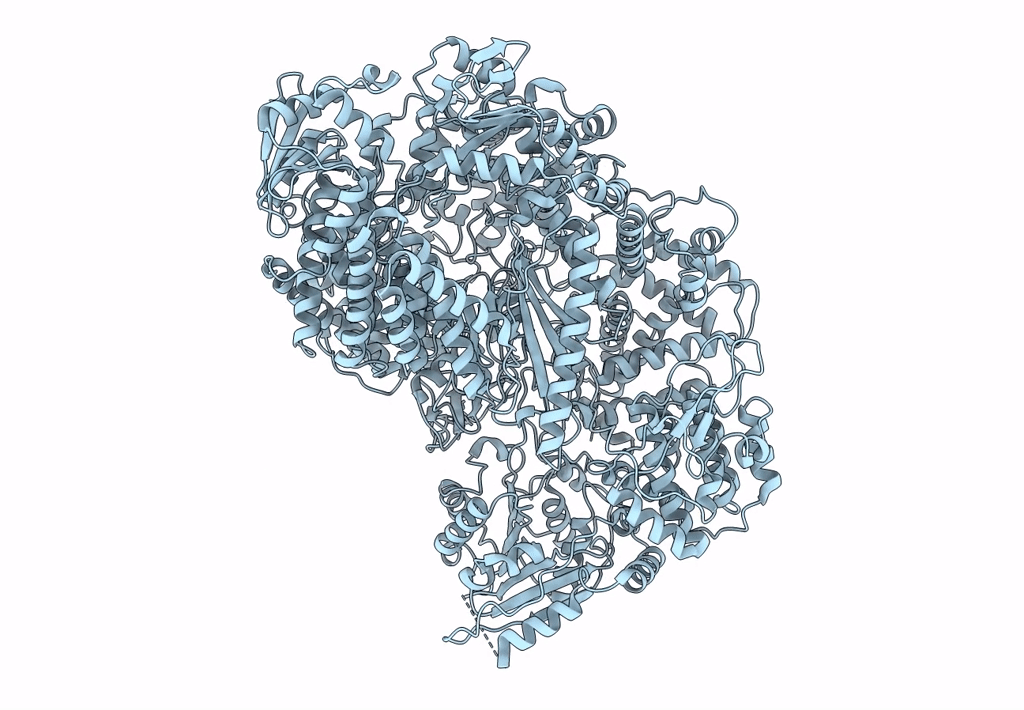
Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy
Biological Source:
Source Organism:
VESICULAR STOMATITIS VIRUS (Taxon ID: 11276)
Host Organism:
Method Details:
Experimental Method:
Resolution:
3.80 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE