Deposition Date
2015-05-20
Release Date
2015-09-23
Last Version Date
2024-11-06
Entry Detail
PDB ID:
4ZXL
Keywords:
Title:
CpOGA D298N in complex with Drosophila HCF -derived Thr-O-GlcNAc peptide
Biological Source:
Source Organism(s):
Clostridium perfringens (strain ATCC 13124 / NCTC 8237 / Type A) (Taxon ID: 195103)
Drosophila melanogaster (Taxon ID: 7227)
Drosophila melanogaster (Taxon ID: 7227)
Expression System(s):
Method Details:
Experimental Method:
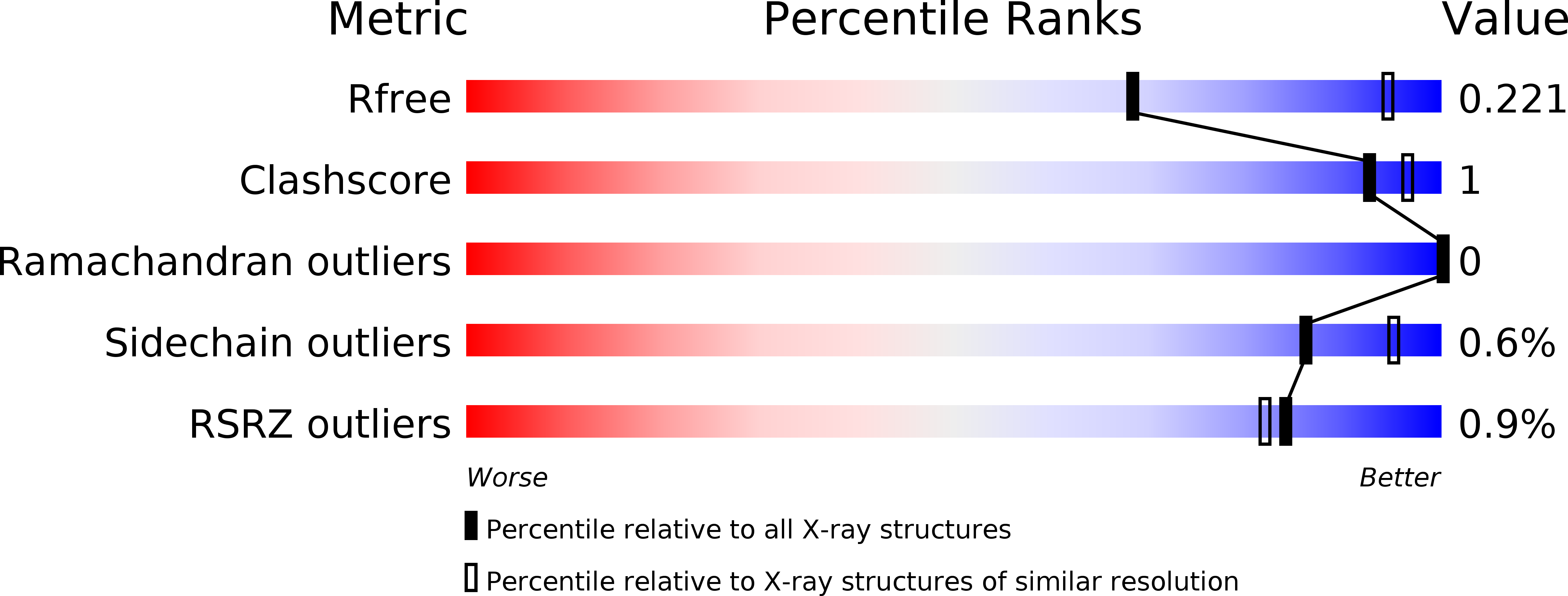
Resolution:
2.60 Å
R-Value Free:
0.22
R-Value Work:
0.17
R-Value Observed:
0.17
Space Group:
P 61