Deposition Date
2015-05-05
Release Date
2015-09-02
Last Version Date
2023-11-15
Entry Detail
PDB ID:
4ZO1
Keywords:
Title:
Crystal Structure of the T3-bound TR-beta Ligand-binding Domain in complex with RXR-alpha
Biological Source:
Source Organism:
Homo sapiens (Taxon ID: 9606)
Host Organism:
Method Details:
Experimental Method:
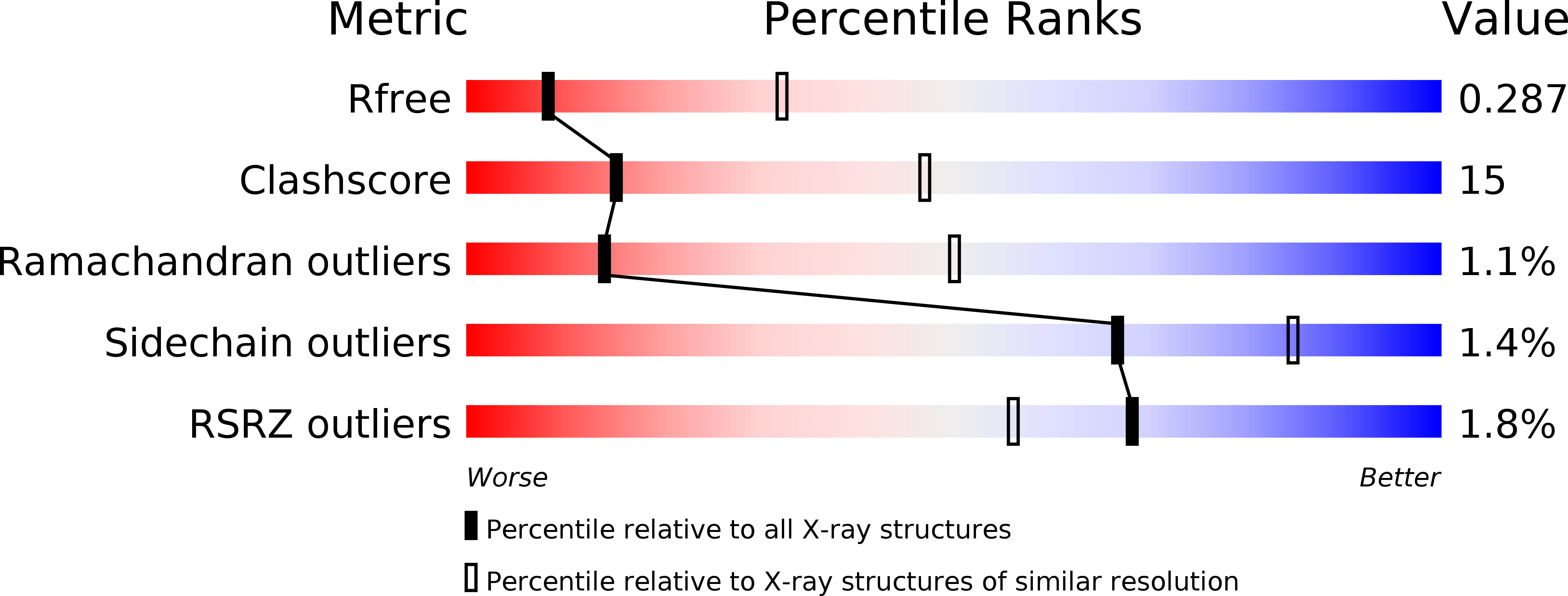
Resolution:
3.22 Å
R-Value Free:
0.28
R-Value Work:
0.23
R-Value Observed:
0.23
Space Group:
P 31 2 1