Deposition Date
2015-04-10
Release Date
2015-05-06
Last Version Date
2023-09-27
Entry Detail
PDB ID:
4Z9L
Keywords:
Title:
THE STRUCTURE OF JNK3 IN COMPLEX WITH AN IMIDAZOLE-PYRIMIDINE INHIBITOR
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.10 Å
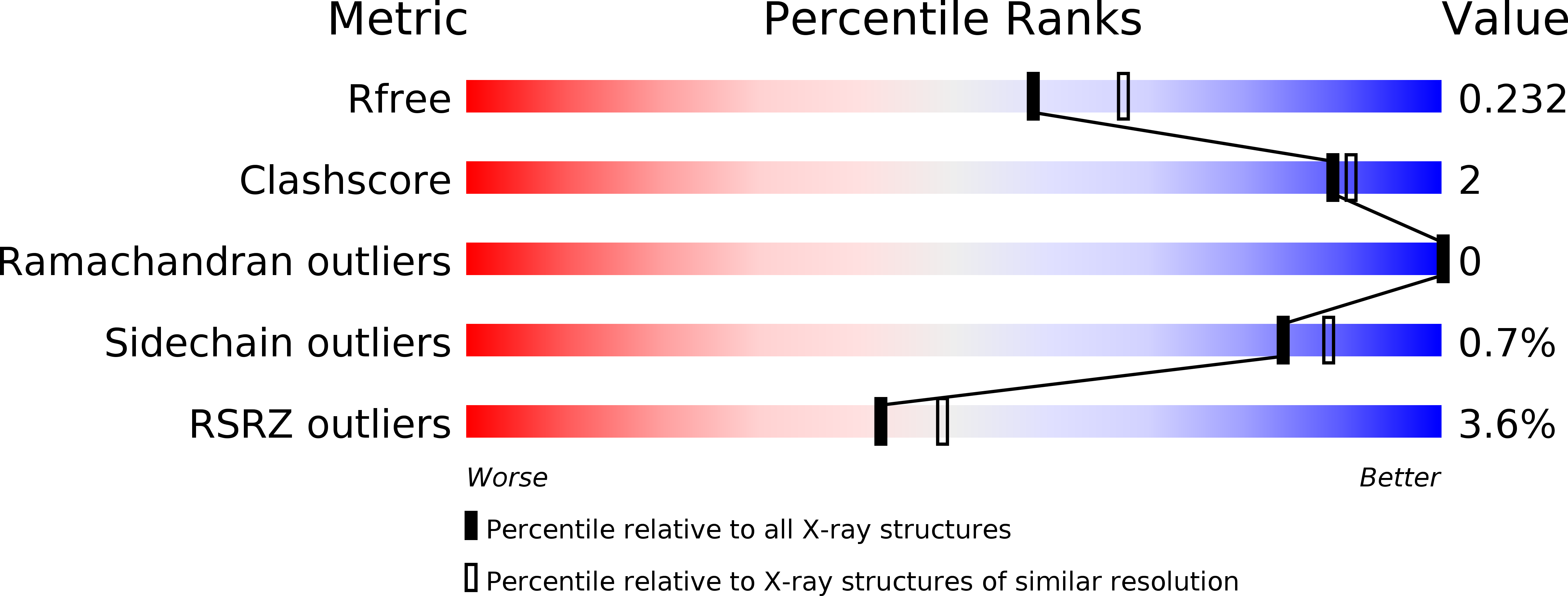
R-Value Free:
0.22
R-Value Work:
0.19
R-Value Observed:
0.20
Space Group:
P 21 21 21