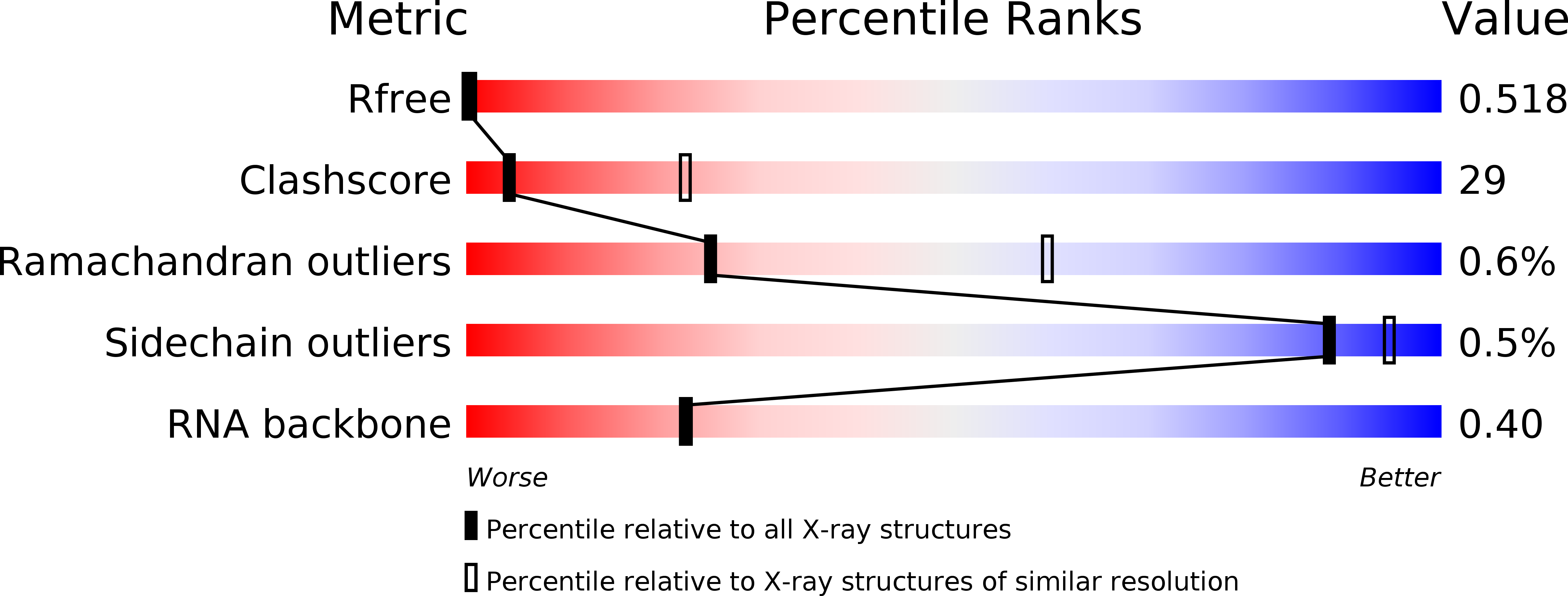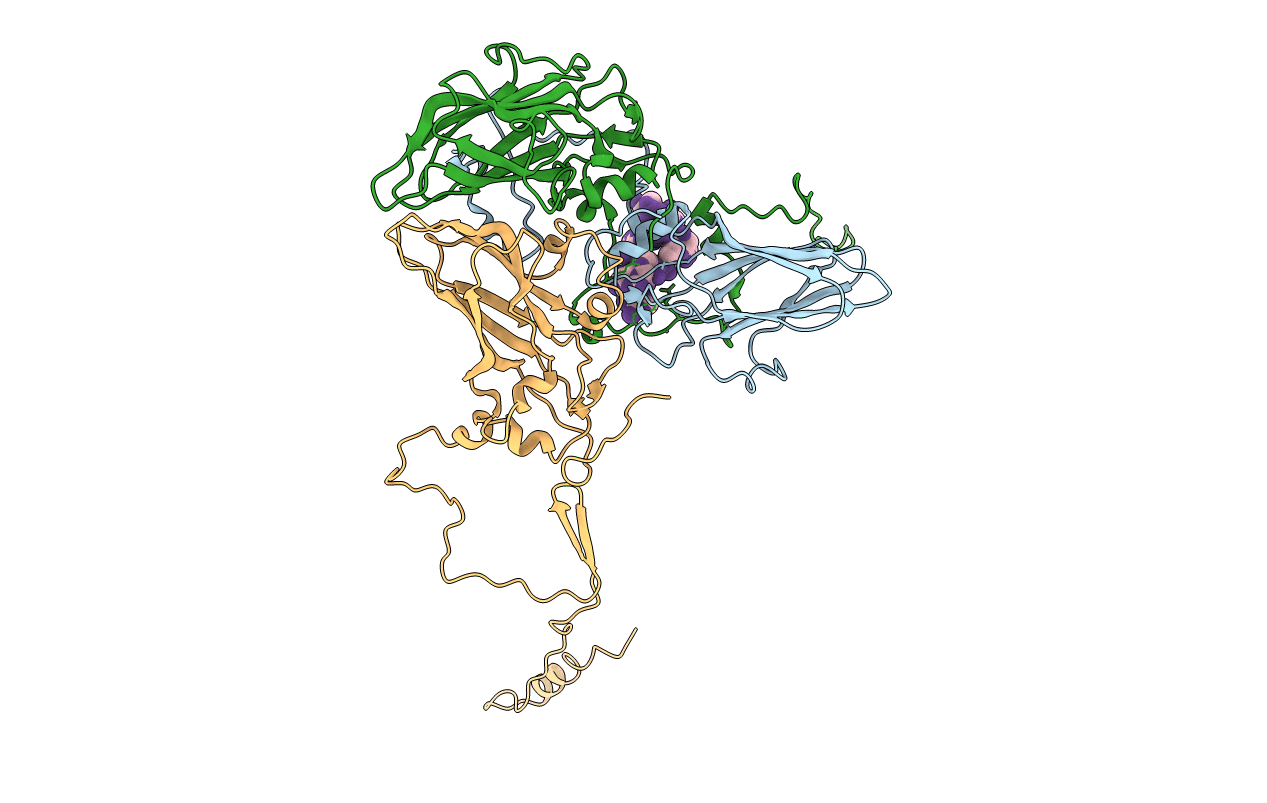
Deposition Date
2015-04-09
Release Date
2015-11-18
Last Version Date
2024-01-10
Entry Detail
Biological Source:
Source Organism(s):
Human parechovirus 1 (strain Harris) (Taxon ID: 103911)
Expression System(s):
Method Details: