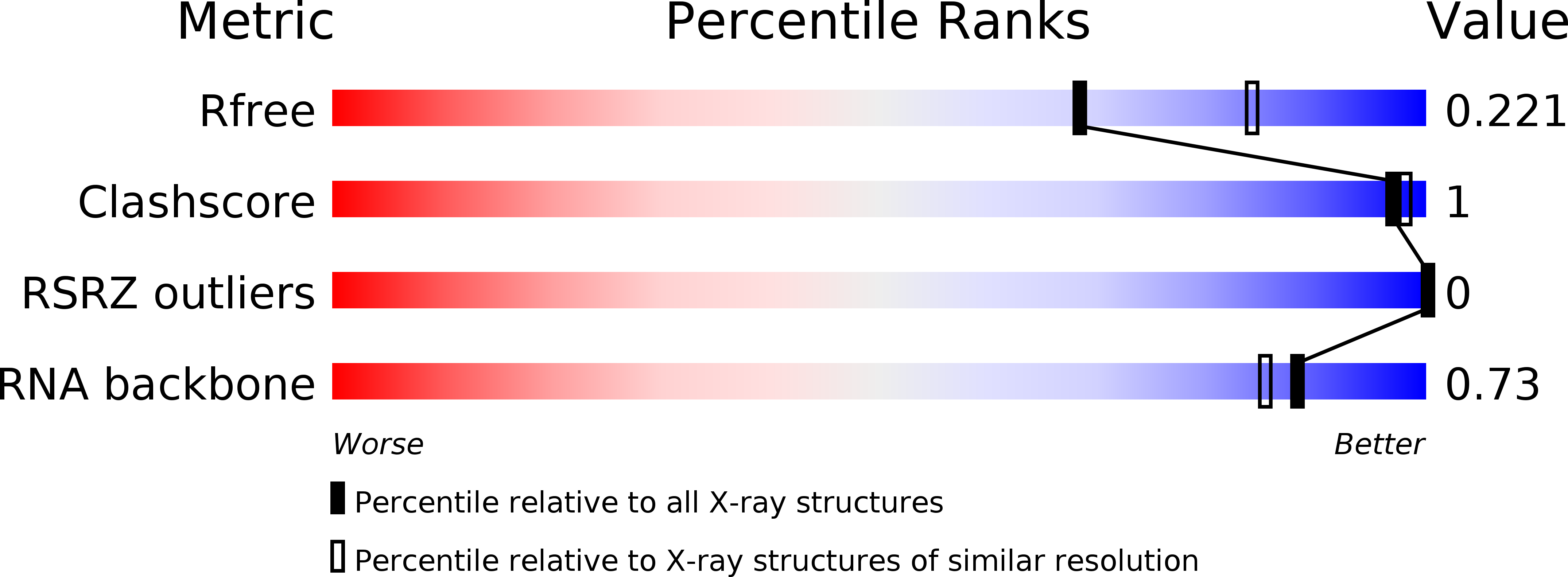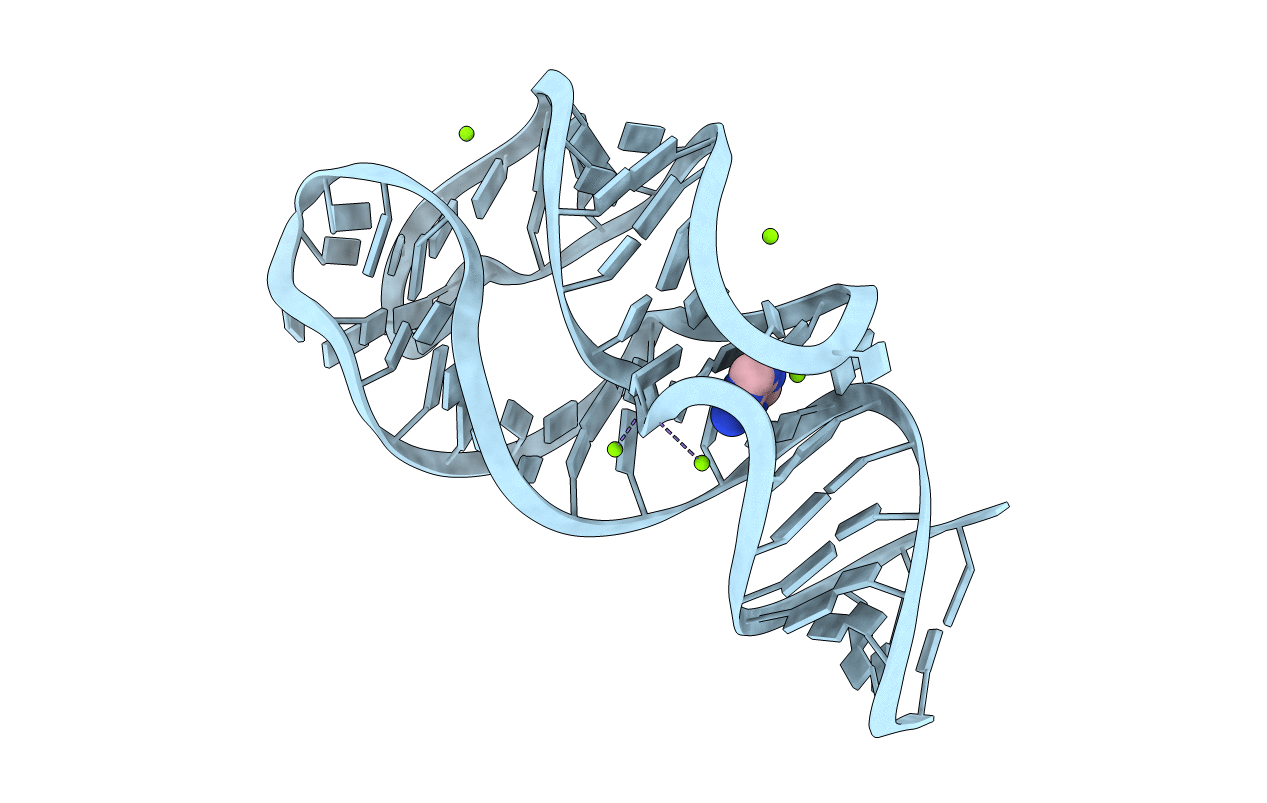
Deposition Date
2015-01-16
Release Date
2015-05-06
Last Version Date
2023-09-27
Entry Detail
PDB ID:
4XNR
Keywords:
Title:
Vibrio Vulnificus Adenine Riboswitch Aptamer Domain, Synthesized by Position-selective Labeling of RNA (PLOR), in Complex with Adenine
Biological Source:
Source Organism:
Vibrio vulnificus (Taxon ID: 672)
Method Details:
Experimental Method:
Resolution:
2.21 Å
R-Value Free:
0.22
R-Value Work:
0.20
R-Value Observed:
0.21
Space Group:
P 21 21 2