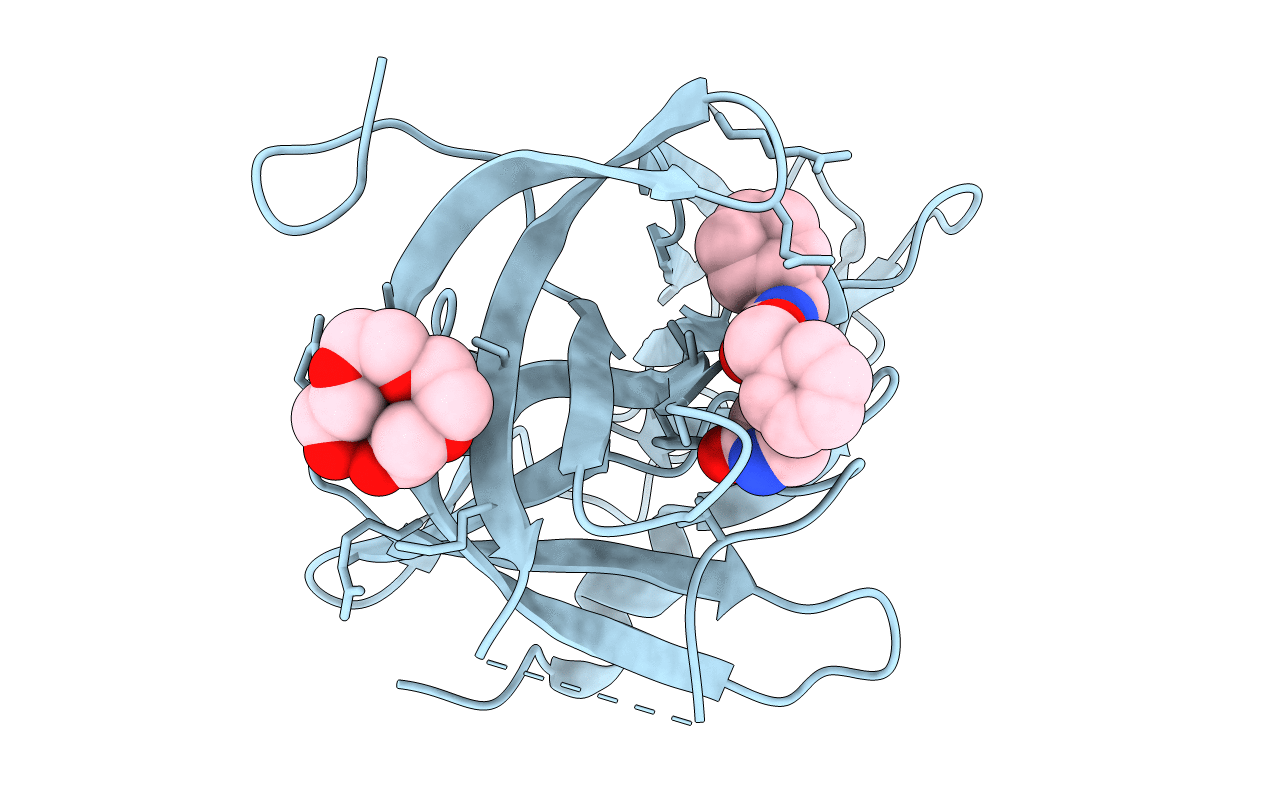
Deposition Date
2014-12-16
Release Date
2015-03-25
Last Version Date
2024-10-23
Entry Detail
PDB ID:
4XBC
Keywords:
Title:
1.60 A resolution structure of Norovirus 3CL protease complex with a covalently bound dipeptidyl inhibitor (1R,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid (Hexagonal Form)
Biological Source:
Source Organism(s):
Expression System(s):
Method Details:
Experimental Method:
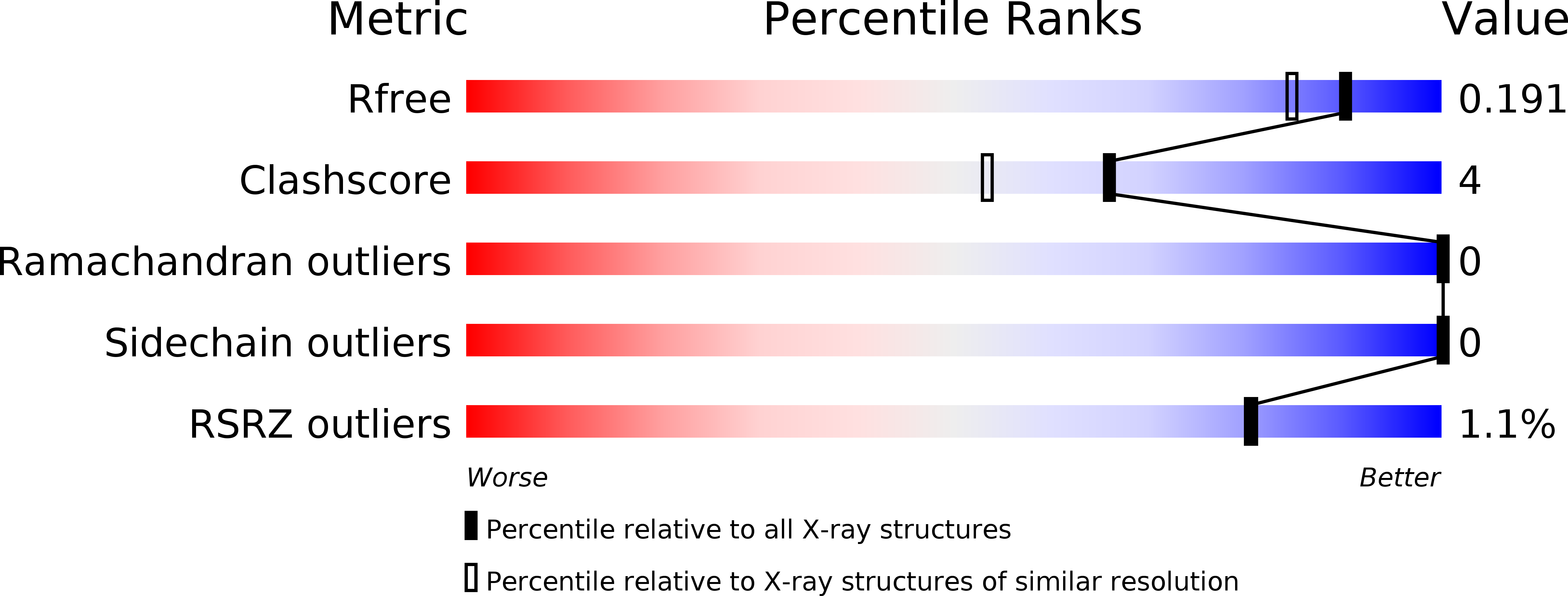
Resolution:
1.60 Å
R-Value Free:
0.17
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
P 65 2 2