Deposition Date
2014-12-09
Release Date
2015-03-04
Last Version Date
2024-02-28
Entry Detail
PDB ID:
4X7V
Keywords:
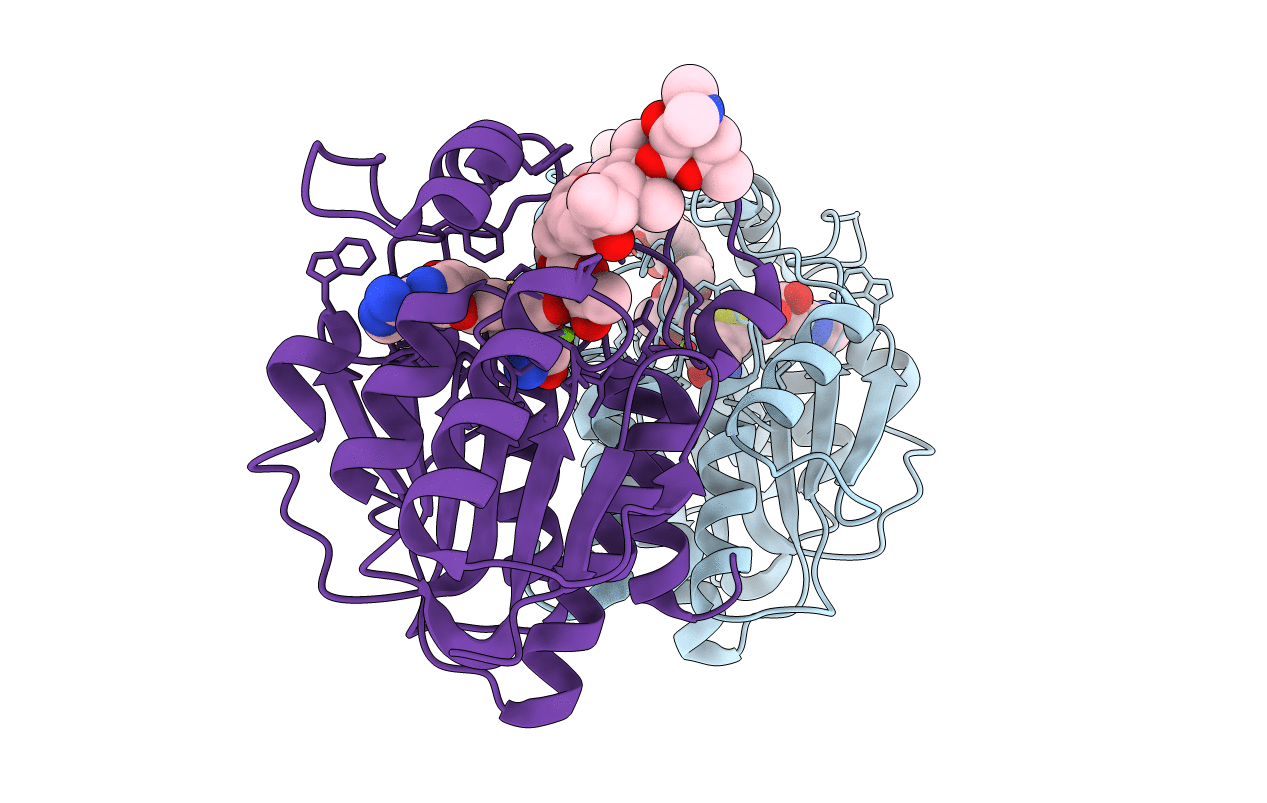
Title:
MycF mycinamicin III 3'-O-methyltransferase (E35Q, E139A variant) in complex with Mg, SAH and mycinamicin IV (product)
Biological Source:
Source Organism(s):
Micromonospora griseorubida (Taxon ID: 28040)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
1.45 Å
R-Value Free:
0.17
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
P 21 21 21


