Deposition Date
2014-12-02
Release Date
2015-03-11
Last Version Date
2024-01-10
Entry Detail
PDB ID:
4X4C
Keywords:
Title:
RADIATION DAMAGE TO THE NUCLEOPROTEIN COMPLEX C.Esp1396I: DOSE (DWD) 6.2 MGy
Biological Source:
Source Organism(s):
Enterobacter sp. RFL1396 (Taxon ID: 211595)
synthetic construct (Taxon ID: 32630)
synthetic construct (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
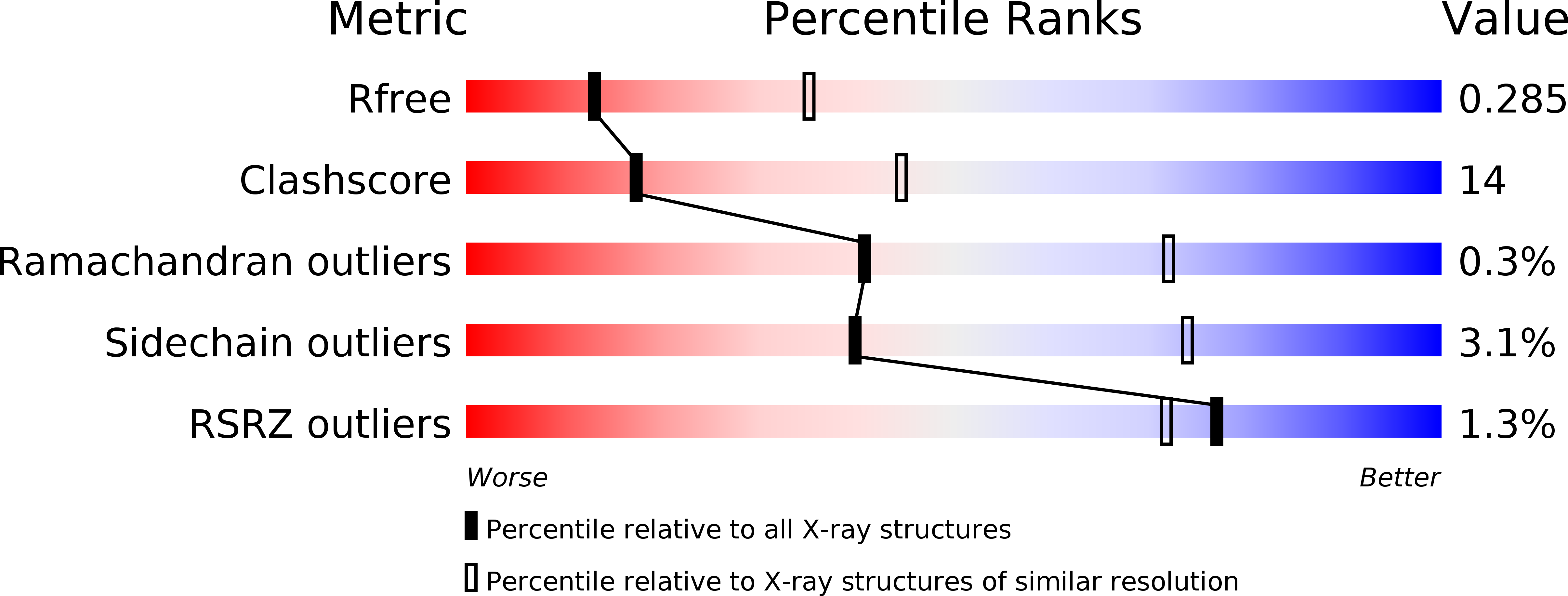
Resolution:
2.80 Å
R-Value Free:
0.27
R-Value Work:
0.23
R-Value Observed:
0.23
Space Group:
P 65