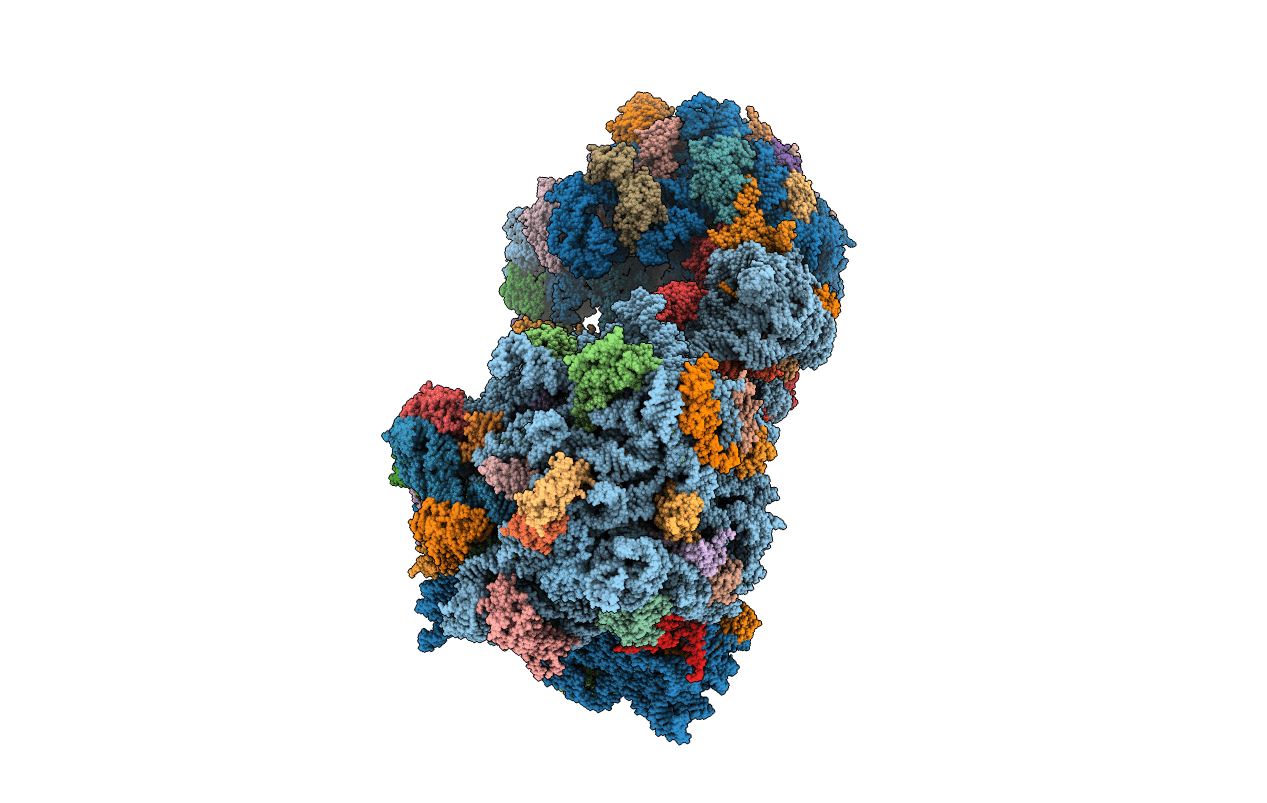
Deposition Date
2014-10-22
Release Date
2015-06-10
Last Version Date
2024-11-06
Entry Detail
PDB ID:
4WQR
Keywords:
Title:
Complex of 70S ribosome with tRNA-Phe and mRNA with C-A mismatch in the first position in the A-site.
Biological Source:
Source Organism(s):
Thermus thermophilus HB8 (Taxon ID: 300852)
Escherichia coli K-12 (Taxon ID: 83333)
Escherichia coli K-12 (Taxon ID: 83333)
Method Details:
Experimental Method:
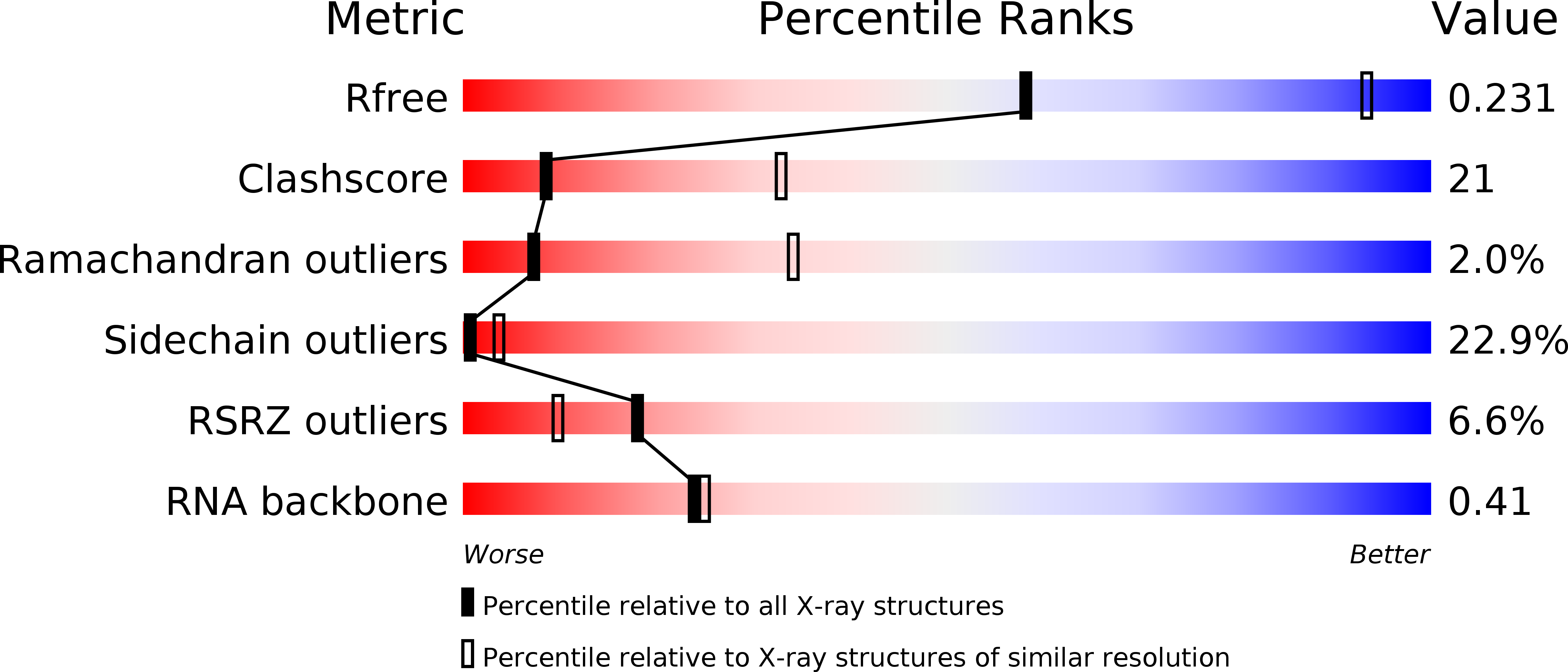
Resolution:
3.15 Å
R-Value Free:
0.23
R-Value Work:
0.18
Space Group:
P 21 21 21