Deposition Date
2017-09-25
Release Date
2017-12-27
Last Version Date
2024-11-13
Entry Detail
PDB ID:
4W2R
Keywords:
Title:
Structure of Hs/AcPRC2 in complex with 5,8-dichloro-2-[(4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-7-[(R)-methoxy(oxetan-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one
Biological Source:
Source Organism(s):
Anolis carolinensis (Taxon ID: 28377)
Homo sapiens (Taxon ID: 9606)
Homo sapiens (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
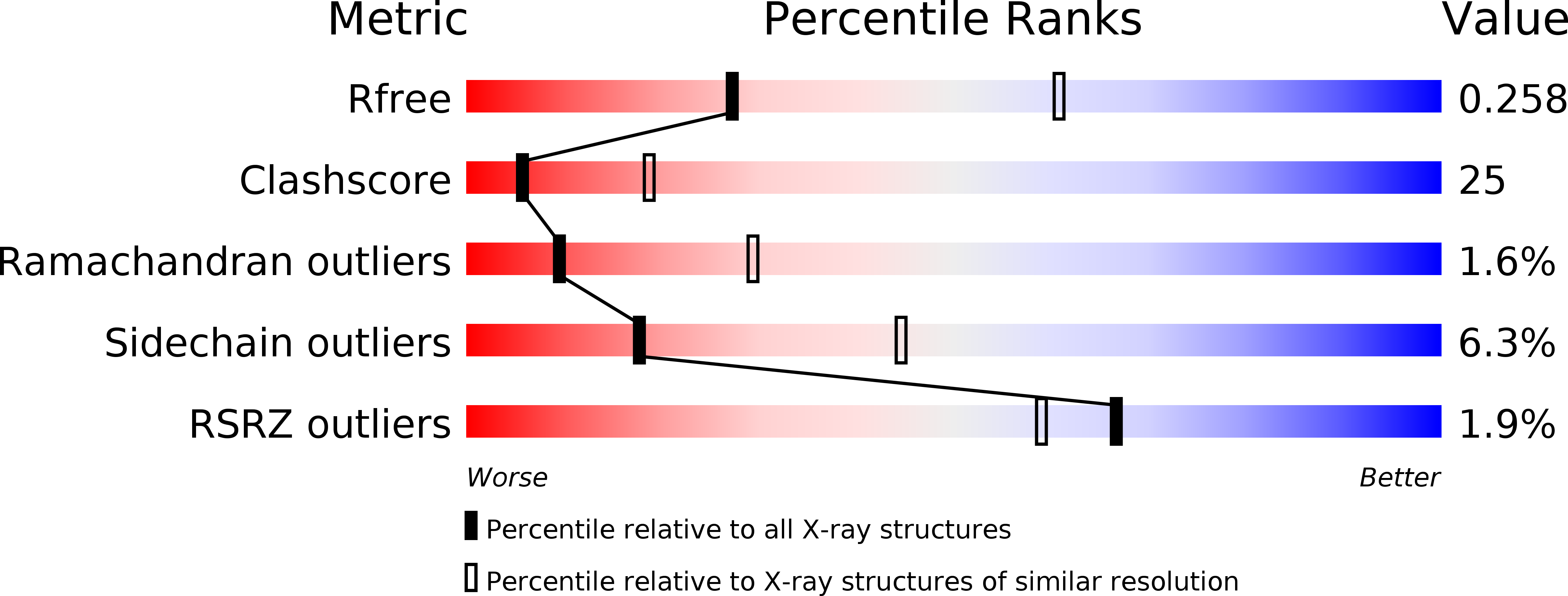
Resolution:
2.81 Å
R-Value Free:
0.26
R-Value Work:
0.21
R-Value Observed:
0.22
Space Group:
P 1 21 1