Deposition Date
2010-12-13
Release Date
2014-07-09
Last Version Date
2024-11-20
Entry Detail
PDB ID:
4V83
Keywords:
Title:
Crystal structure of a complex containing domain 3 from the PSIV IGR IRES RNA bound to the 70S ribosome.
Biological Source:
Source Organism(s):
Thermus thermophilus (Taxon ID: 262724)
Method Details:
Experimental Method:
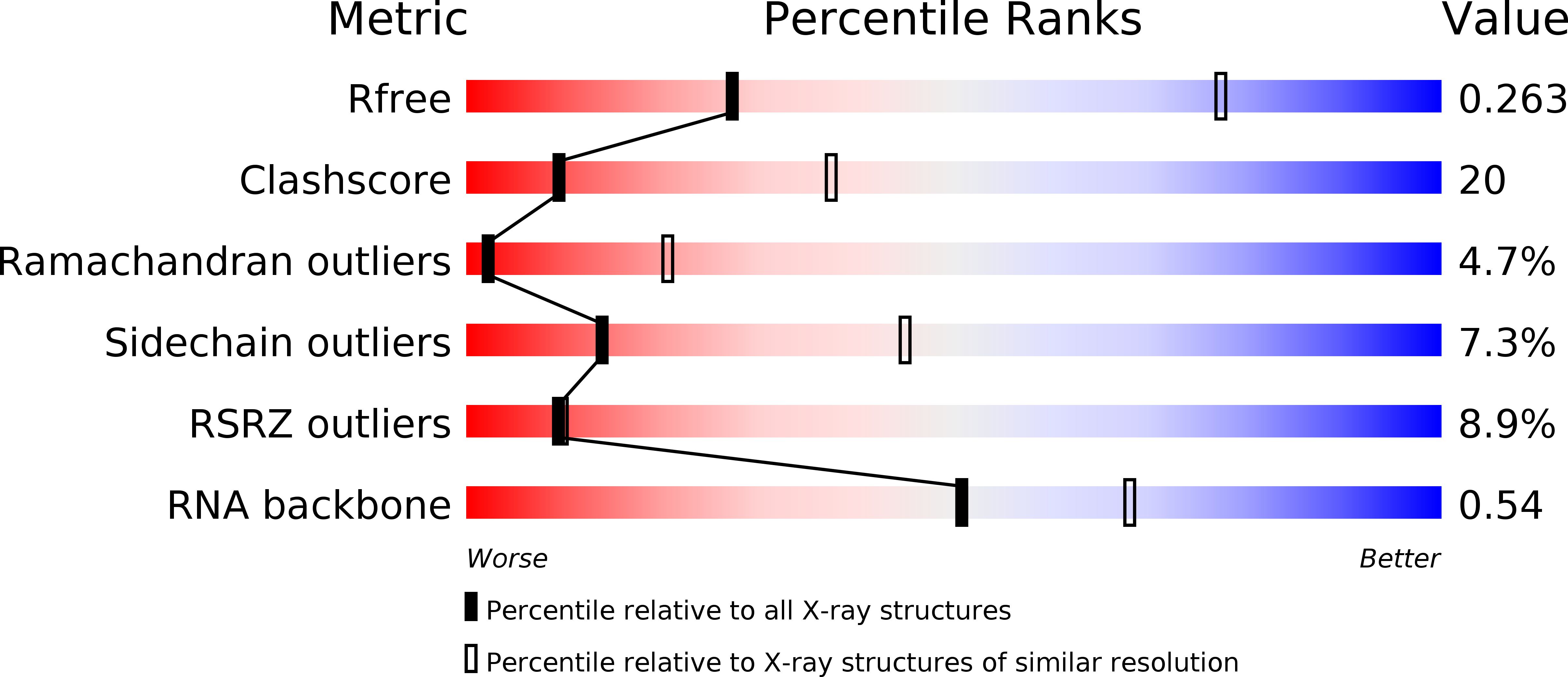
Resolution:
3.50 Å
R-Value Free:
0.26
R-Value Work:
0.23
R-Value Observed:
0.23
Space Group:
P 21 21 21