Deposition Date
2004-01-06
Release Date
2014-07-09
Last Version Date
2024-02-28
Entry Detail
PDB ID:
4V4B
Keywords:
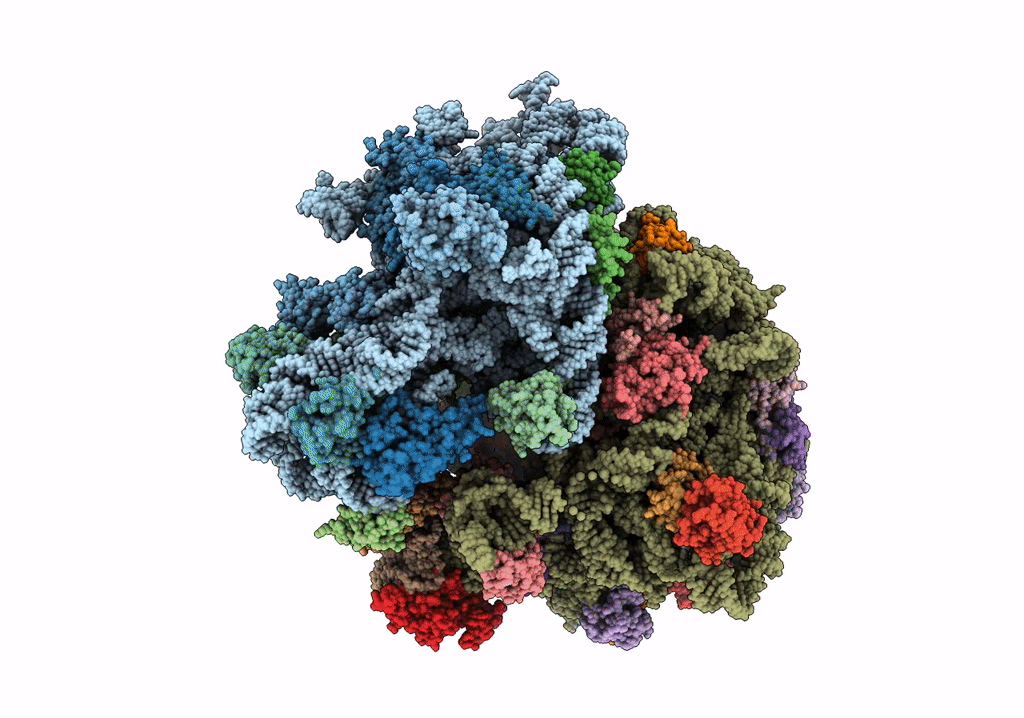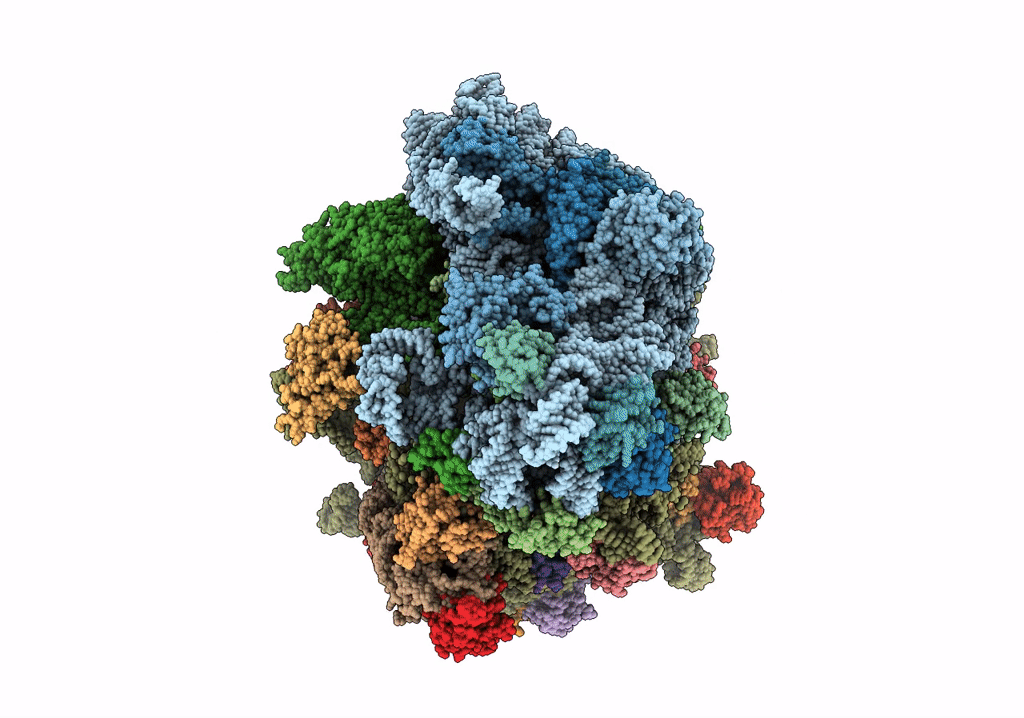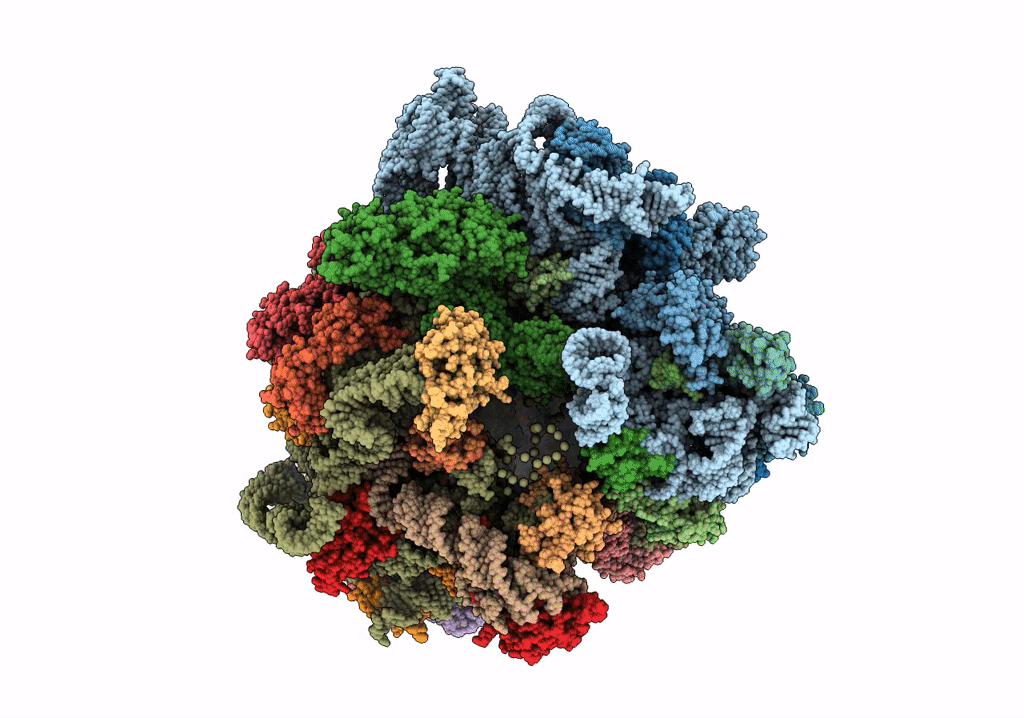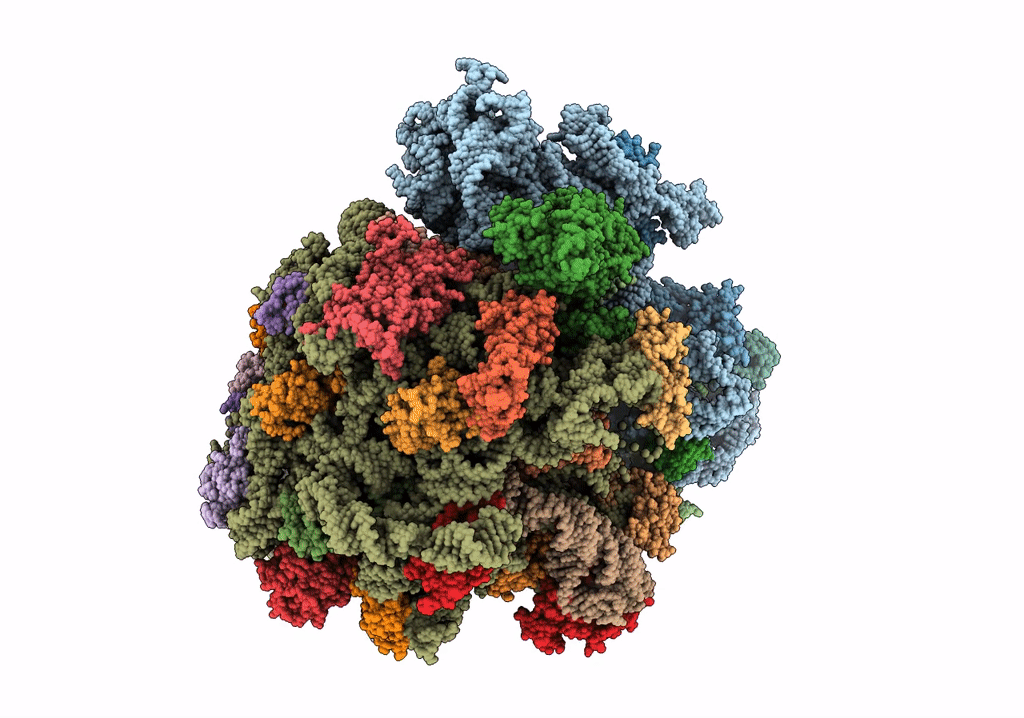
Title:
Structure of the ribosomal 80S-eEF2-sordarin complex from yeast obtained by docking atomic models for RNA and protein components into a 11.7 A cryo-EM map.
Biological Source:
Source Organism(s):
Saccharomyces cerevisiae (Taxon ID: 4932)
Method Details:
Experimental Method:
Resolution:
11.70 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


