Deposition Date
2014-06-24
Release Date
2015-08-12
Last Version Date
2024-01-10
Entry Detail
PDB ID:
4UQM
Keywords:
Title:
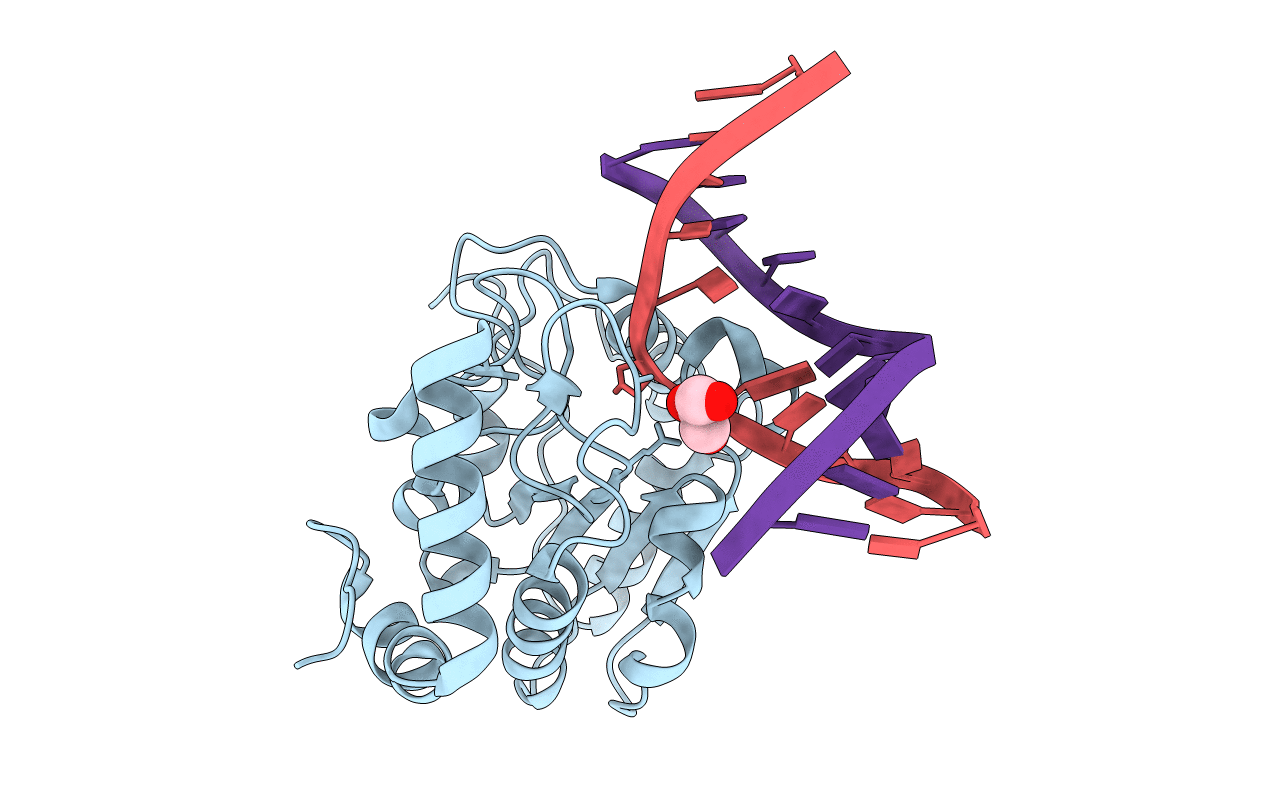
Crystal structure determination of uracil-DNA N-glycosylase (UNG) from Deinococcus radiodurans in complex with DNA - new insights into the role of the Leucine-loop for damage recognition and repair
Biological Source:
Source Organism(s):
DEINOCOCCUS RADIODURANS (Taxon ID: 1299)
SYNTHETIC CONSTRUCT (Taxon ID: 32630)
SYNTHETIC CONSTRUCT (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
1.35 Å
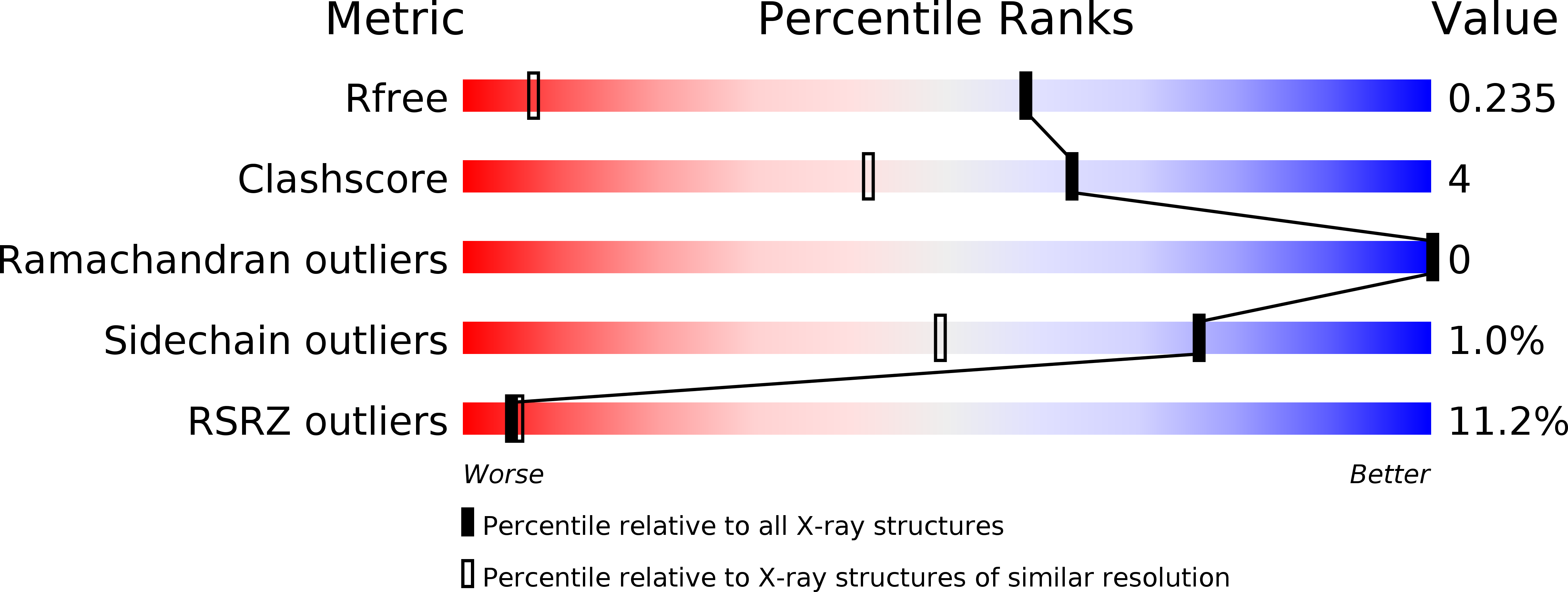
R-Value Free:
0.21
R-Value Work:
0.18
R-Value Observed:
0.18
Space Group:
P 21 21 2