Deposition Date
2015-03-17
Release Date
2016-01-27
Last Version Date
2024-10-09
Entry Detail
PDB ID:
4UFG
Keywords:
Title:
Thrombin in complex with (2R)-2-(benzylsulfonylamino)-N-((1S)-2-((4- carbamimidoylphenyl)methylamino)-1-methyl-2-oxo-ethyl)-N-methyl-3- phenyl-propanamide ethane
Biological Source:
Source Organism(s):
HIRUDO MEDICINALIS (Taxon ID: 6421)
HOMO SAPIENS (Taxon ID: 9606)
HOMO SAPIENS (Taxon ID: 9606)
Method Details:
Experimental Method:
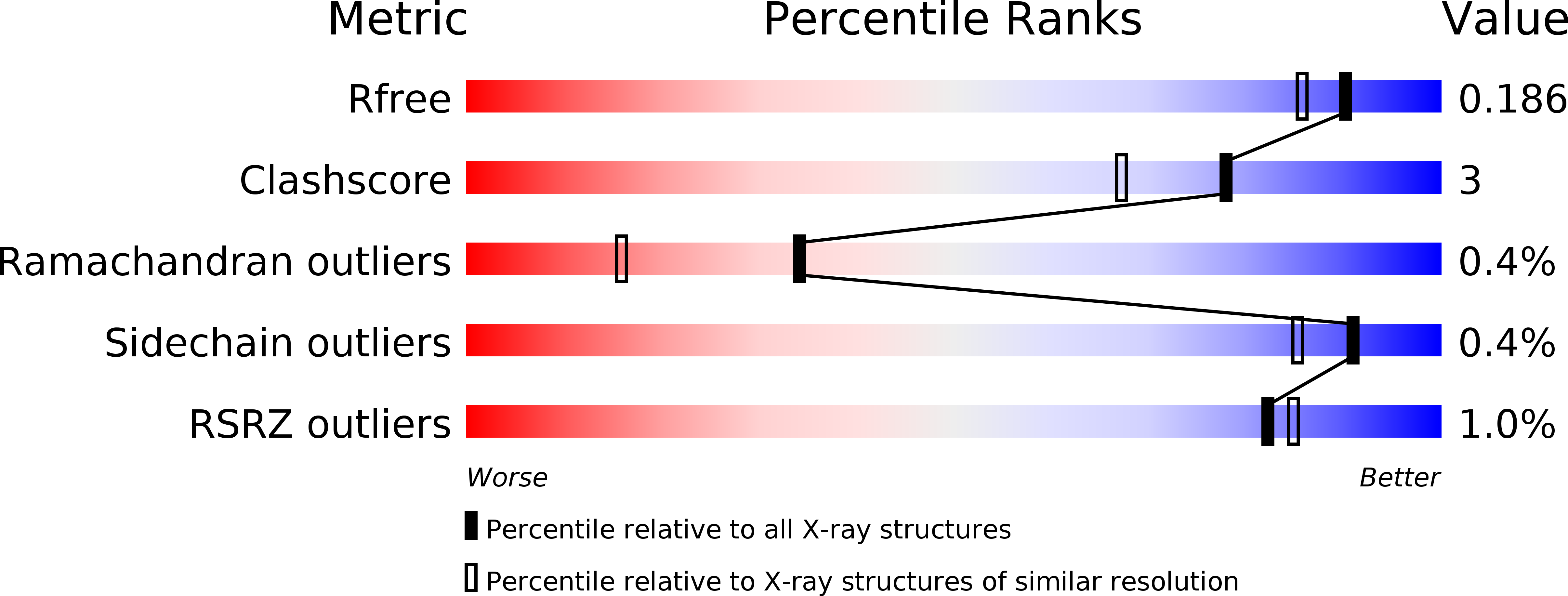
Resolution:
1.65 Å
R-Value Free:
0.18
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
C 1 2 1