Deposition Date
2014-12-15
Release Date
2015-09-09
Last Version Date
2024-05-08
Entry Detail
PDB ID:
4UE5
Keywords:
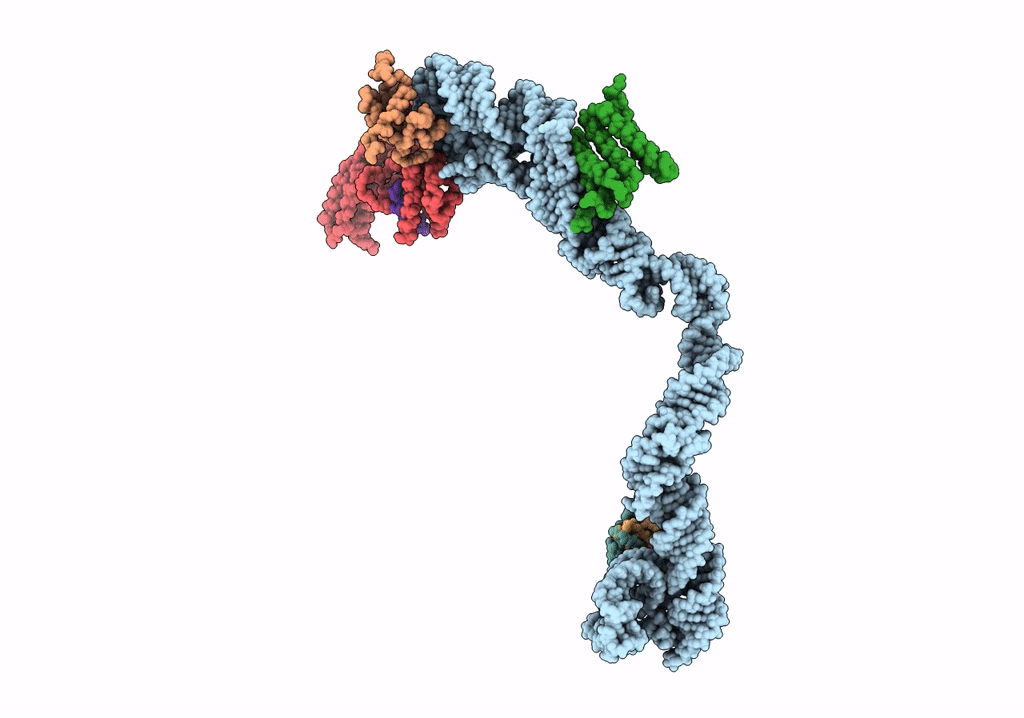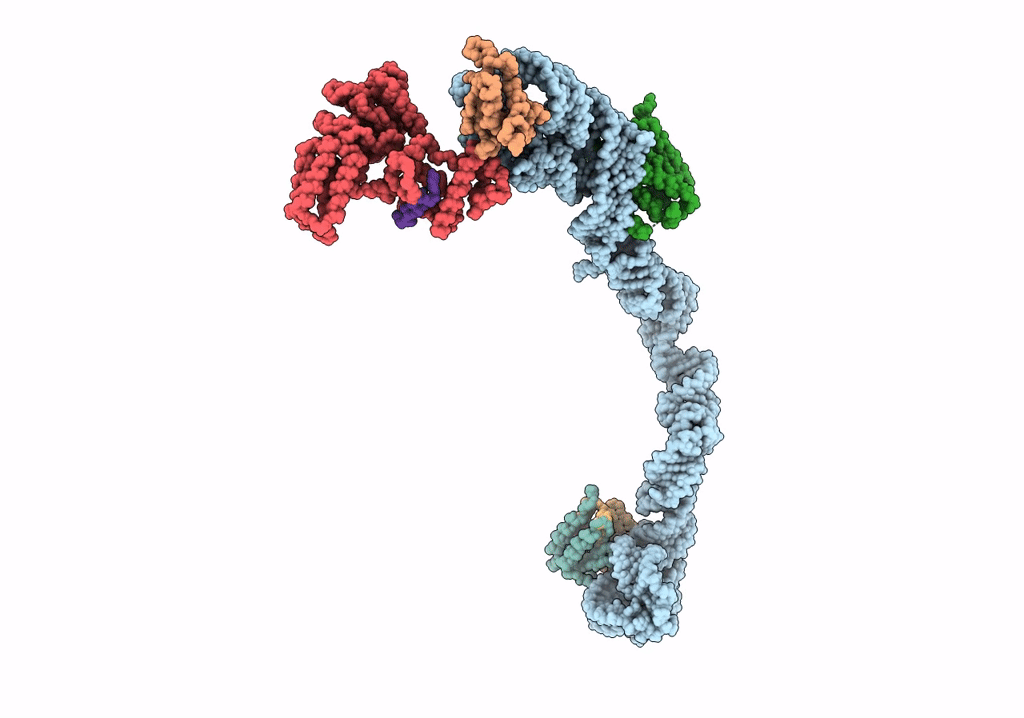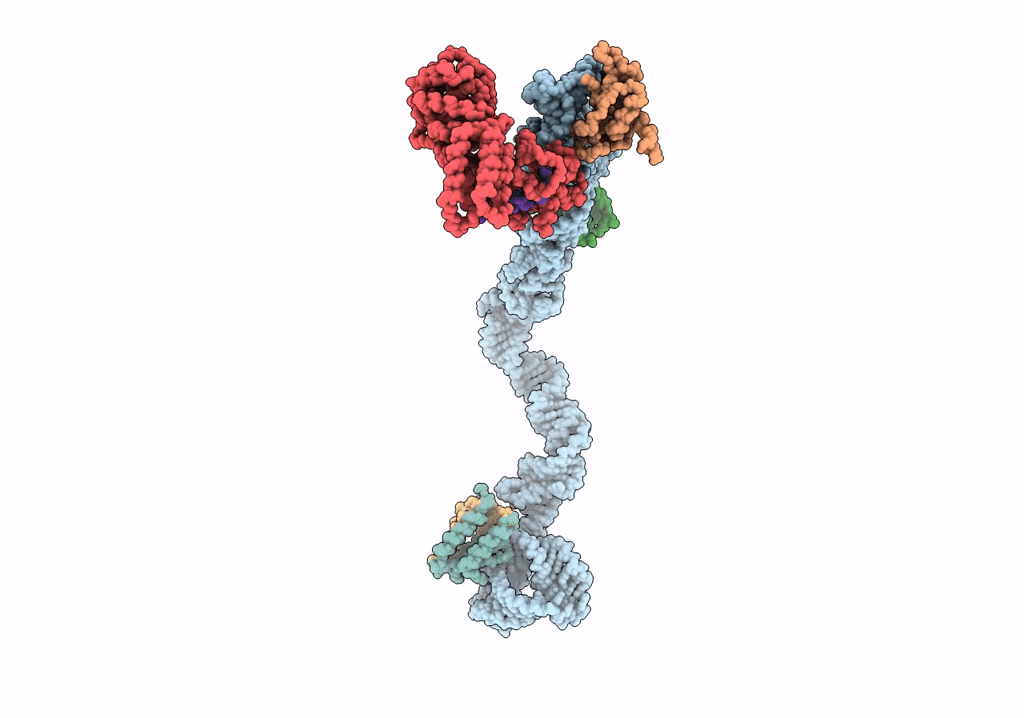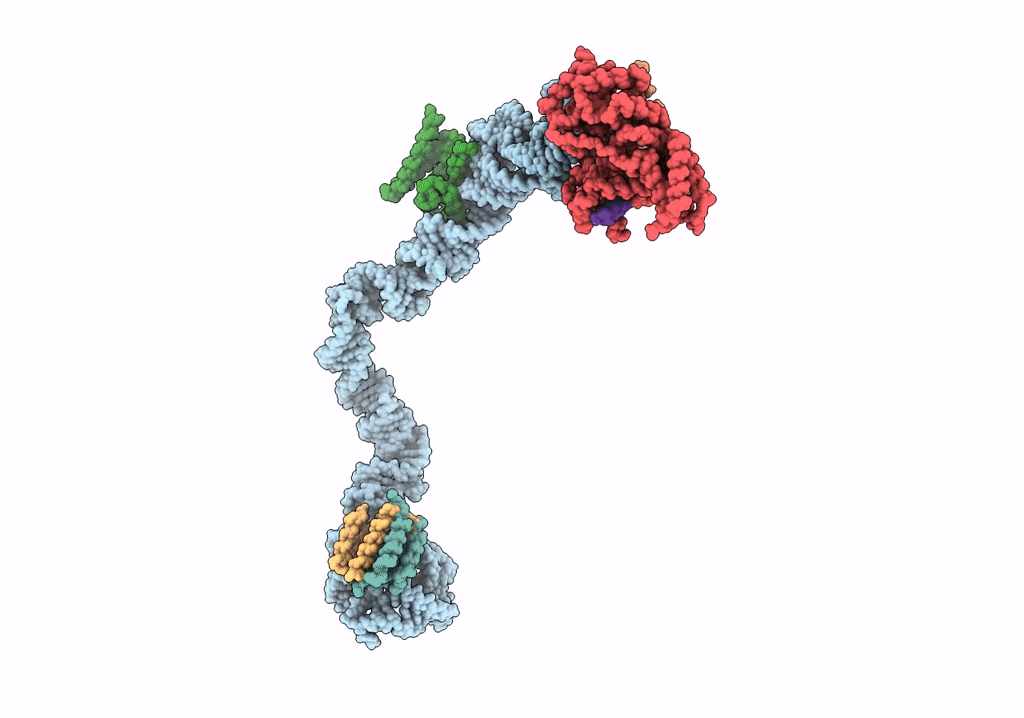
Title:
Structural basis for targeting and elongation arrest of Bacillus signal recognition particle
Biological Source:
Source Organism(s):
CANIS LUPUS FAMILIARIS (Taxon ID: 9615)
Method Details:
Experimental Method:
Resolution:
9.00 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


