Deposition Date
2014-07-11
Release Date
2014-09-10
Last Version Date
2024-10-09
Entry Detail
PDB ID:
4TZV
Keywords:
Title:
Co-crystals of the Ternary Complex Containing a T-box Stem I RNA, its Cognate tRNAGly, and B. subtilis YbxF protein, treated by removing lithium sulfate post crystallization
Biological Source:
Source Organism(s):
Bacillus subtilis (Taxon ID: 1423)
Oceanobacillus iheyensis HTE831 (Taxon ID: 221109)
Oceanobacillus iheyensis HTE831 (Taxon ID: 221109)
Expression System(s):
Method Details:
Experimental Method:
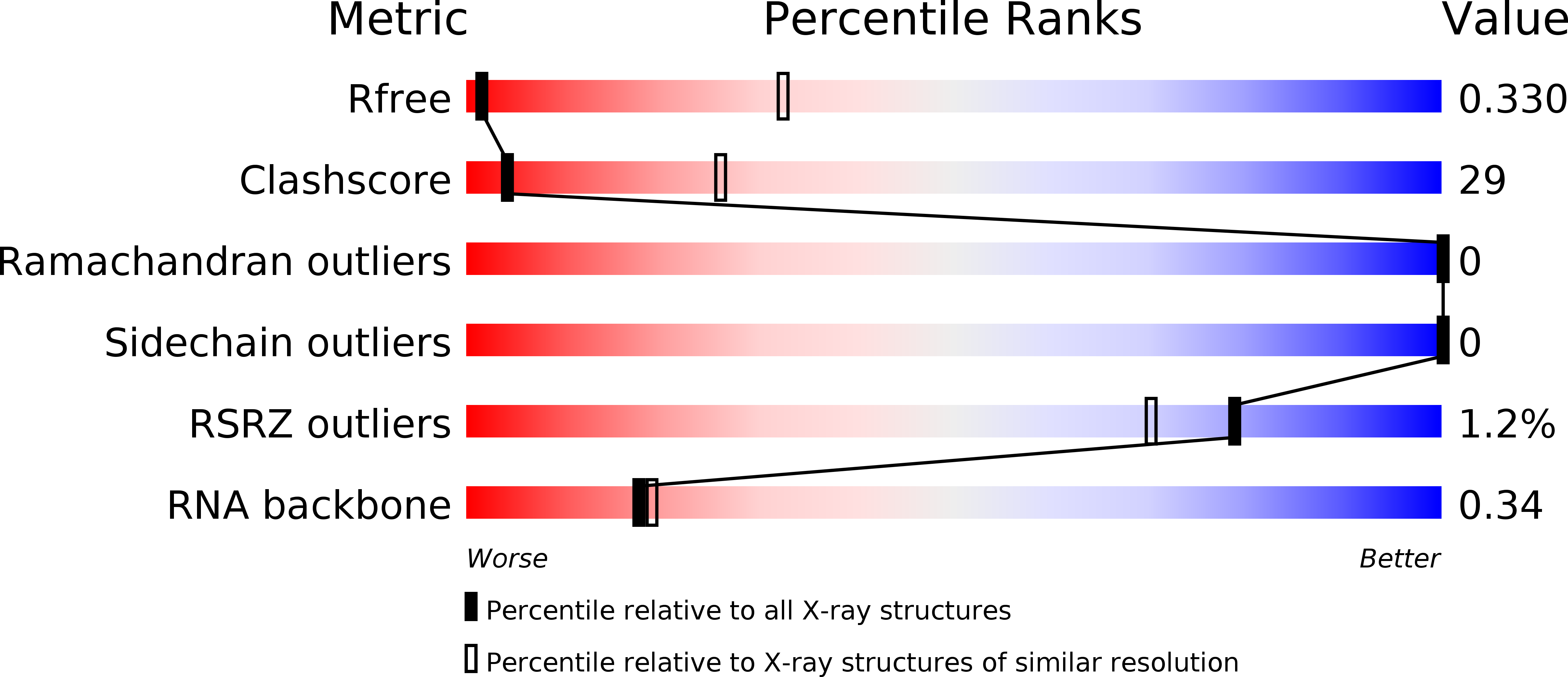
Resolution:
5.03 Å
R-Value Free:
0.32
R-Value Work:
0.27
R-Value Observed:
0.28
Space Group:
P 43 21 2