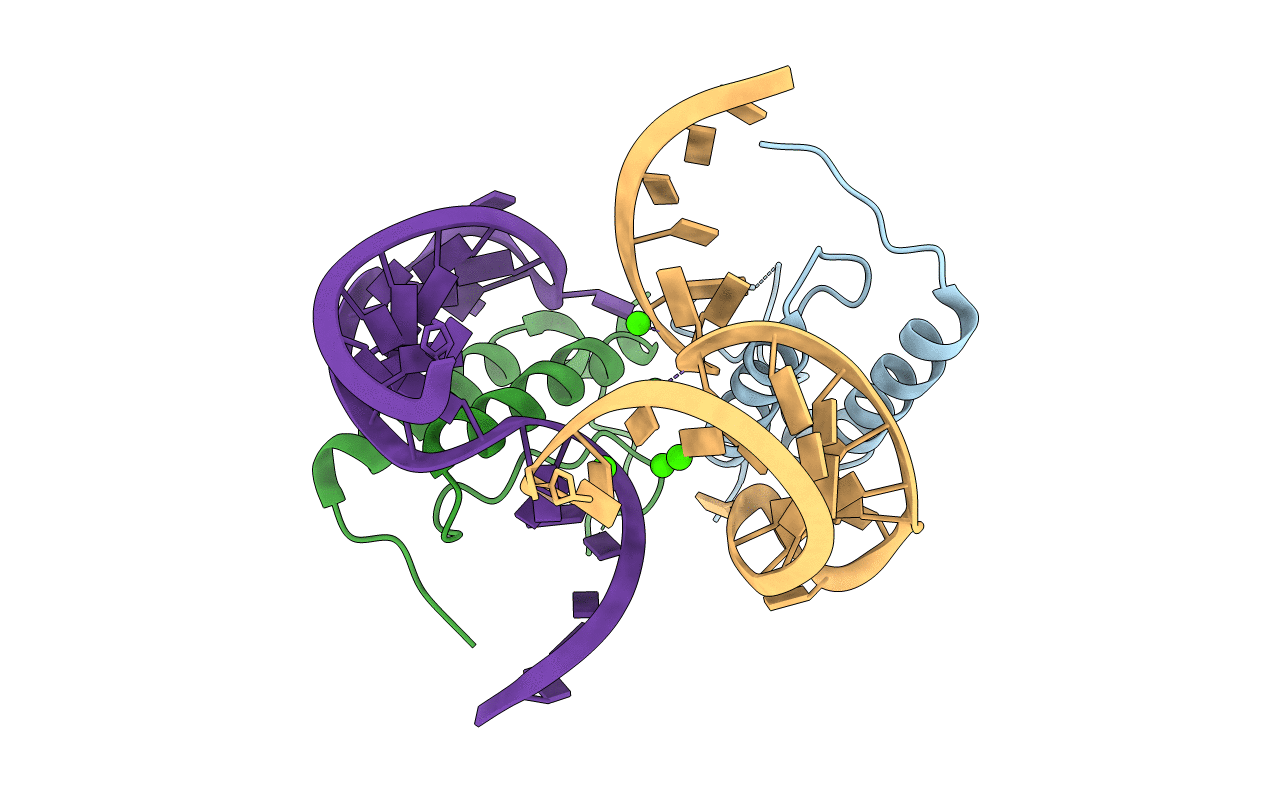
Deposition Date
2014-06-25
Release Date
2014-07-23
Last Version Date
2023-12-27
Entry Detail
PDB ID:
4TUW
Keywords:
Title:
drosophila stem-loop binding protein complexed with histone mRNA stem-loop, phospho mimic of TPNK and C-terminal region
Biological Source:
Source Organism(s):
Drosophila melanogaster (Taxon ID: 7227)
synthetic construct (Taxon ID: 32630)
synthetic construct (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
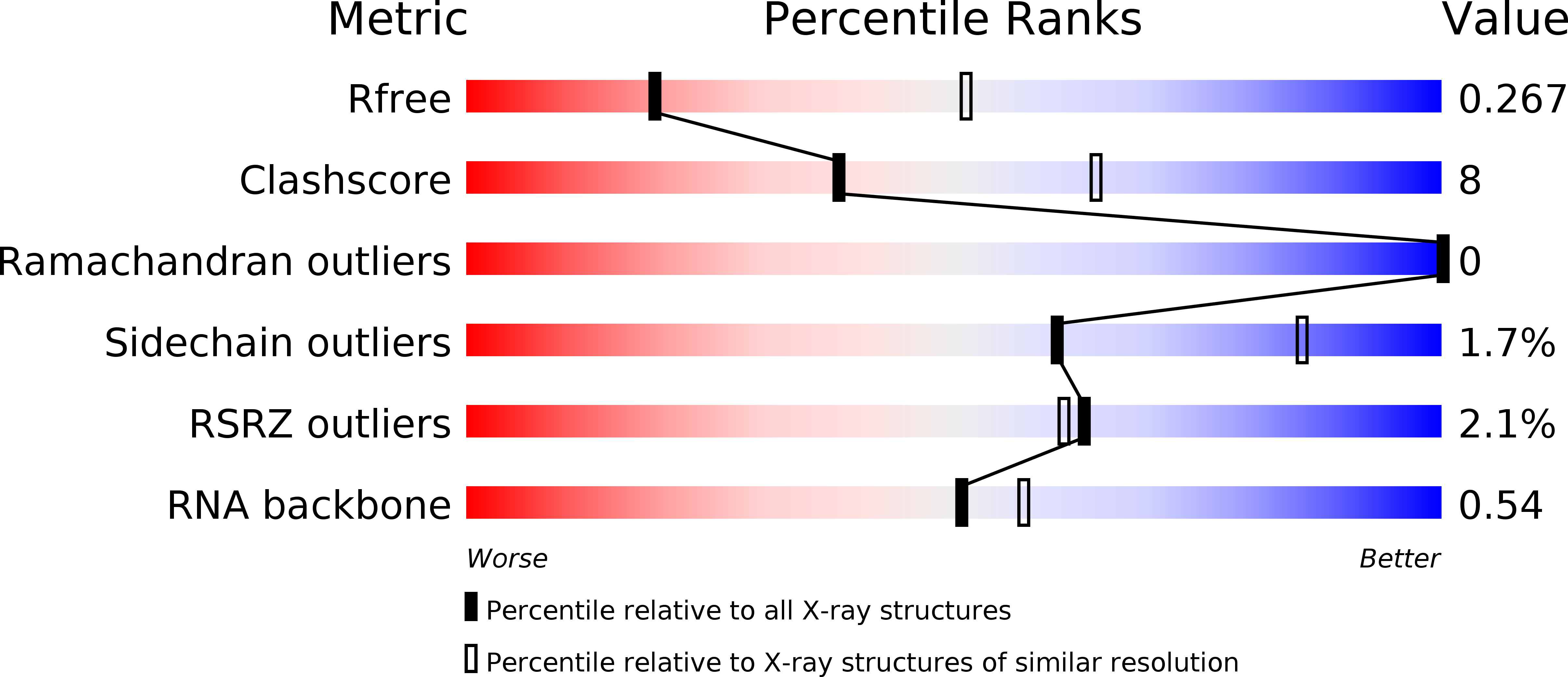
Resolution:
2.90 Å
R-Value Free:
0.26
R-Value Work:
0.21
R-Value Observed:
0.21
Space Group:
P 43 21 2