Deposition Date
2014-10-07
Release Date
2015-08-12
Last Version Date
2024-11-27
Entry Detail
PDB ID:
4RIS
Keywords:
Title:
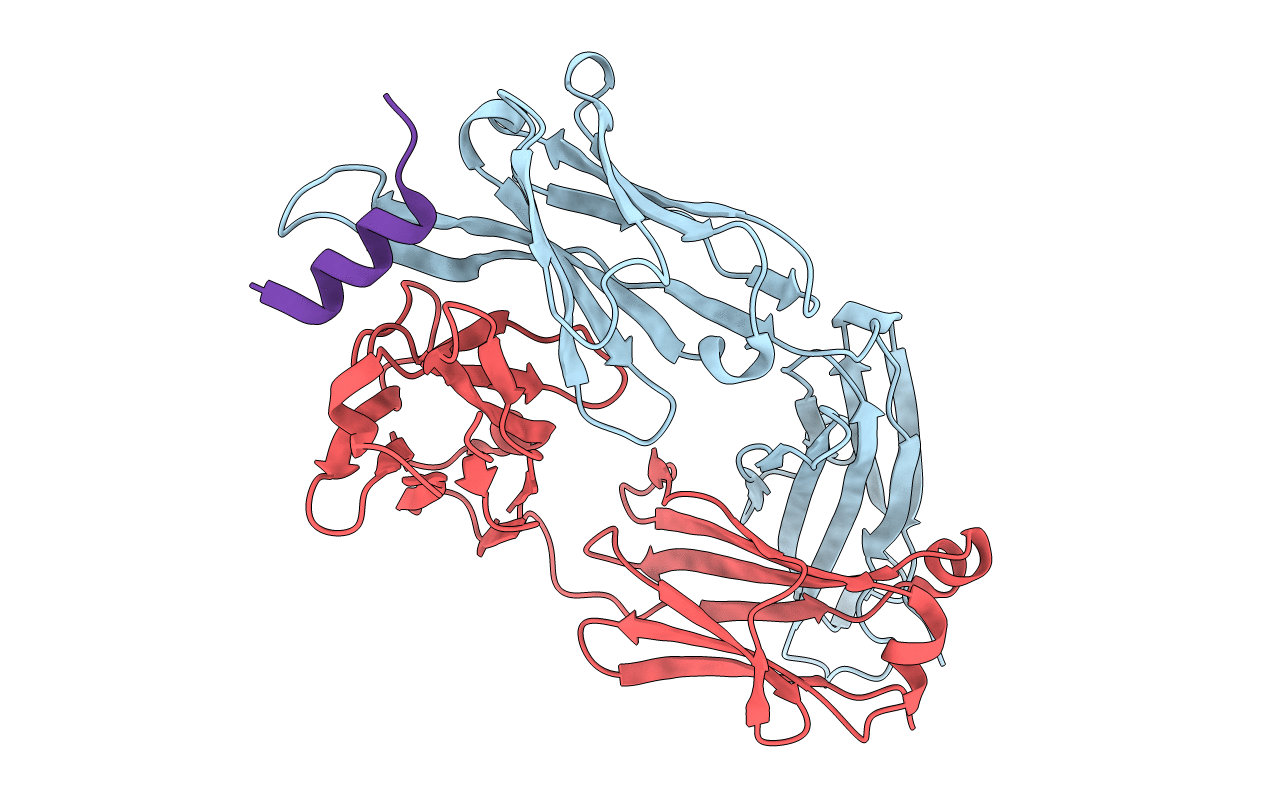
Structural Analysis of the Unmutated Ancestor of the HIV-1 Envelope V2 Region Antibody CH58 Isolated From an RV144 HIV-1 Vaccine Efficacy Trial Vaccinee and Associated with Decreased Transmission Risk
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Human immunodeficiency virus 1 (Taxon ID: 11676)
Human immunodeficiency virus 1 (Taxon ID: 11676)
Expression System(s):
Method Details:
Experimental Method:
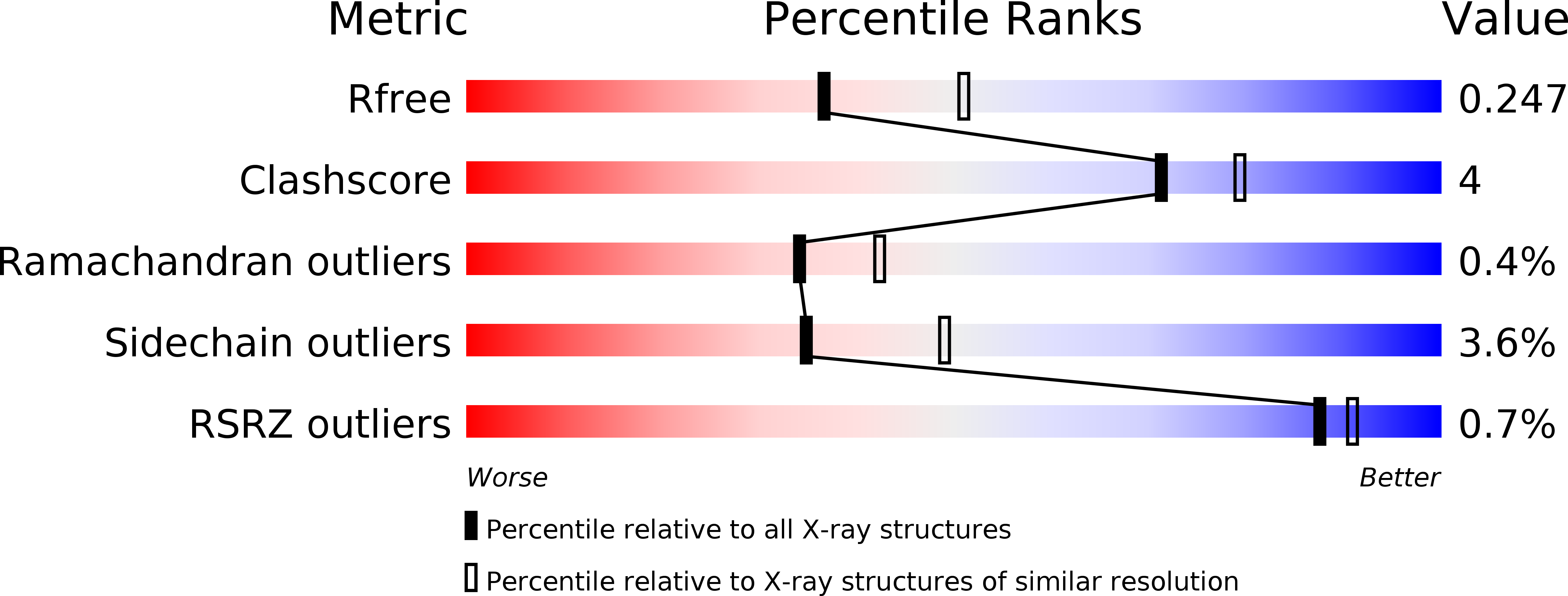
Resolution:
2.30 Å
R-Value Free:
0.24
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
P 1 21 1