Deposition Date
2014-07-22
Release Date
2015-03-18
Last Version Date
2024-02-28
Entry Detail
PDB ID:
4QXX
Keywords:
Title:
Structure of the amyloid forming peptide GNLVS (residues 26-30) from the eosinophil major basic protein (EMBP)
Biological Source:
Source Organism:
Homo sapiens (Taxon ID: 9606)
Method Details:
Experimental Method:
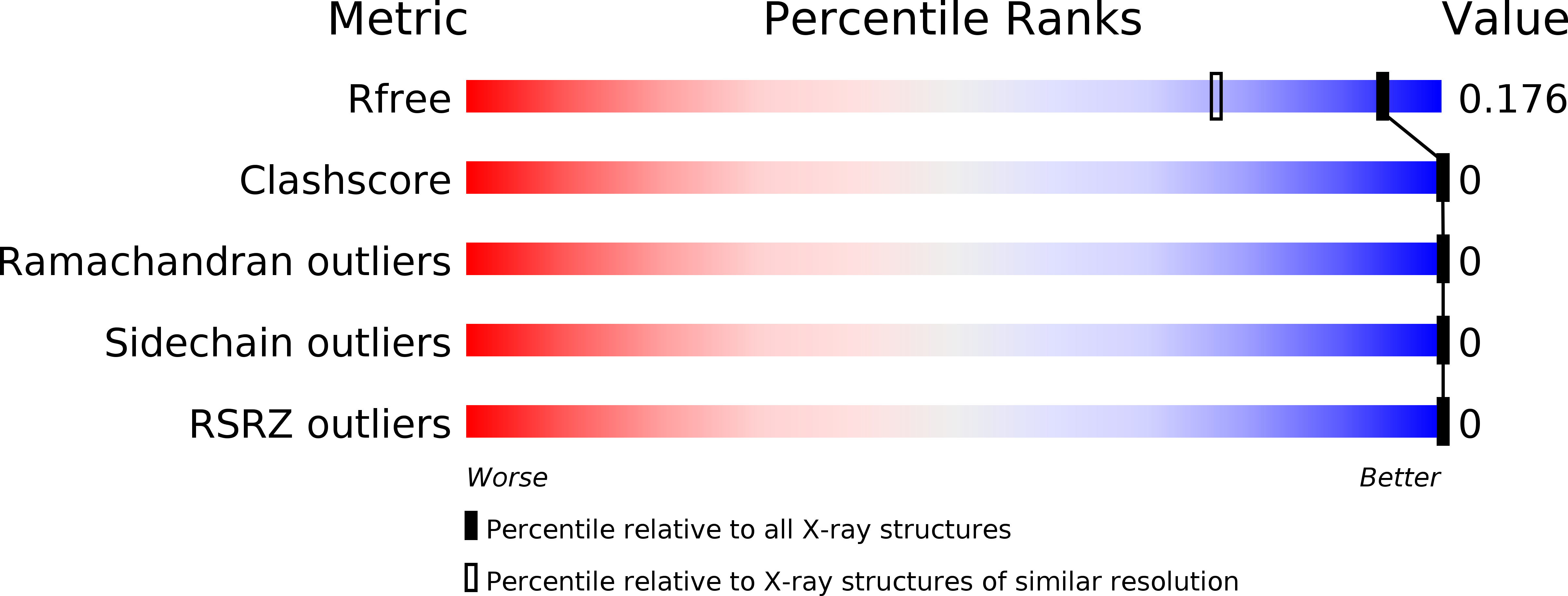
Resolution:
1.45 Å
R-Value Free:
0.19
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
P 2 21 21