Deposition Date
2014-07-07
Release Date
2014-07-23
Last Version Date
2023-09-20
Entry Detail
PDB ID:
4QTE
Keywords:
Title:
Structure of ERK2 in complex with VTX-11e, 4-{2-[(2-CHLORO-4-FLUOROPHENYL)AMINO]-5-METHYLPYRIMIDIN-4-YL}-N-[(1S)-1-(3-CHLOROPHENYL)-2-HYDROXYETHYL]-1H-PYRROLE-2-CARBOXAMIDE
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
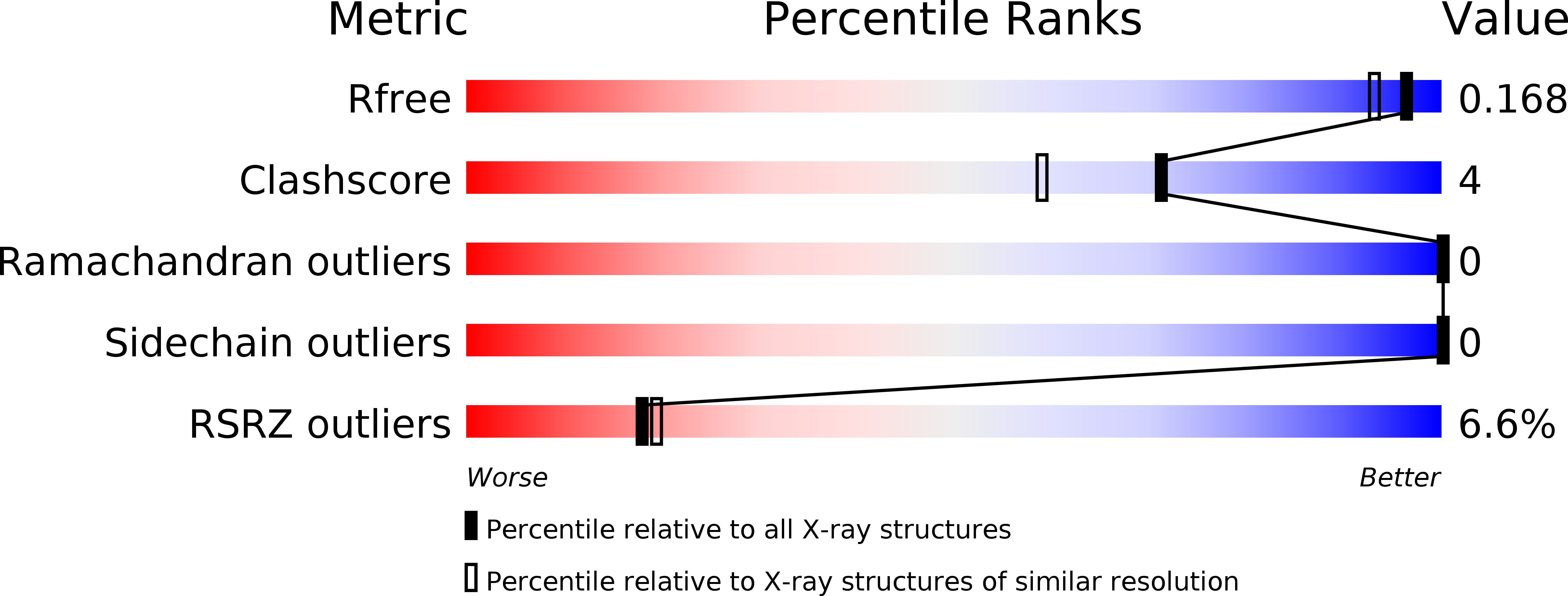
Resolution:
1.50 Å
R-Value Free:
0.16
R-Value Work:
0.14
R-Value Observed:
0.14
Space Group:
P 32 2 1