Deposition Date
2014-05-22
Release Date
2014-06-11
Last Version Date
2024-02-28
Entry Detail
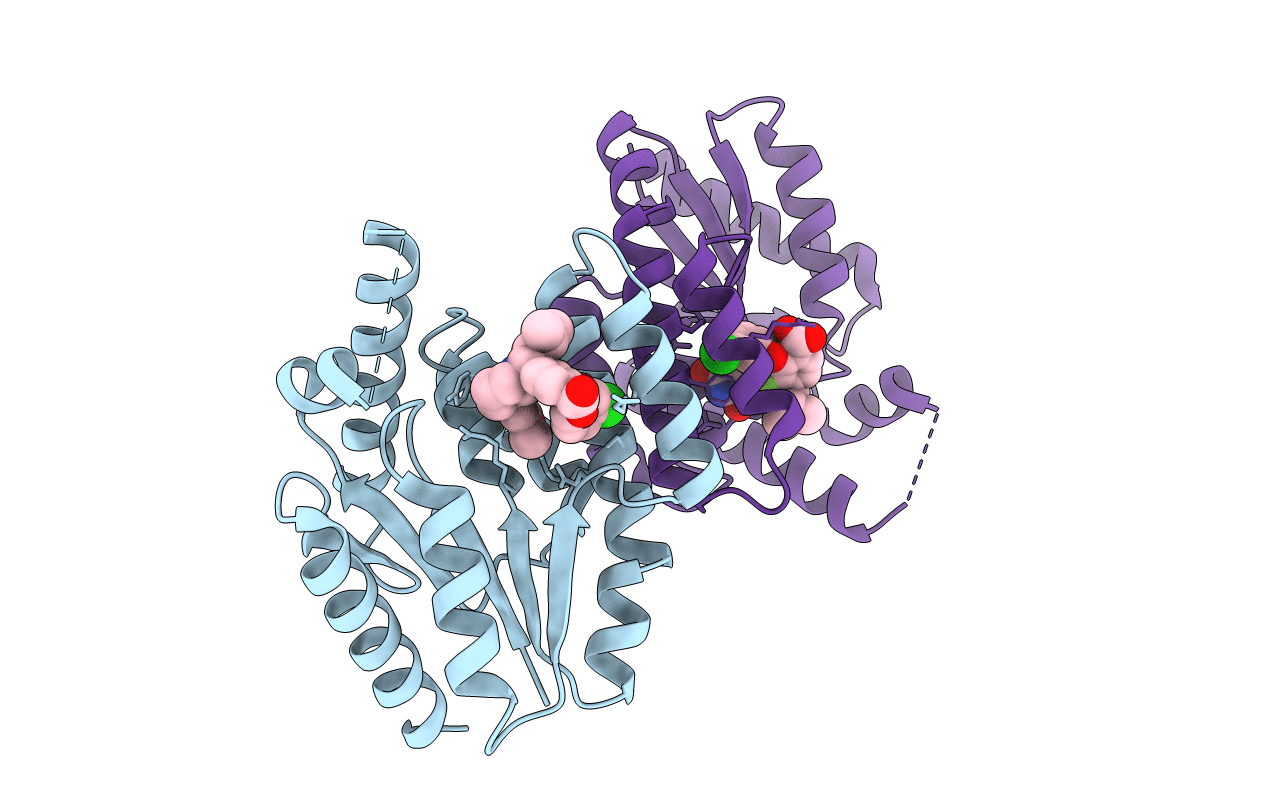
PDB ID:
4QGG
Keywords:
Title:
TMK in complex with compound 46, 2-(3-CHLOROPHENOXY)-3-FLUORO-4-{(1R)-3-METHYL-1-[(3S)-3-(5-METHYL-2,4-DIOXO-3,4-DIHYDROPYRIMIDIN-1(2H)-YL)PIPERIDIN-1-YL]BUTYL}BENZOIC ACID
Biological Source:
Source Organism(s):
Staphylococcus aureus subsp. aureus (Taxon ID: 282458)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
1.62 Å
R-Value Free:
0.22
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
P 1 21 1


