Deposition Date
2014-04-21
Release Date
2014-12-31
Last Version Date
2023-09-20
Entry Detail
PDB ID:
4Q6A
Keywords:
Title:
Staphylococcus aureus V31L, F98Y Mutant Dihydrofolate Reductase Complexed with NADPH
Biological Source:
Source Organism:
Staphylococcus aureus MUF168 (Taxon ID: 1435480)
Host Organism:
Method Details:
Experimental Method:
Resolution:
2.10 Å
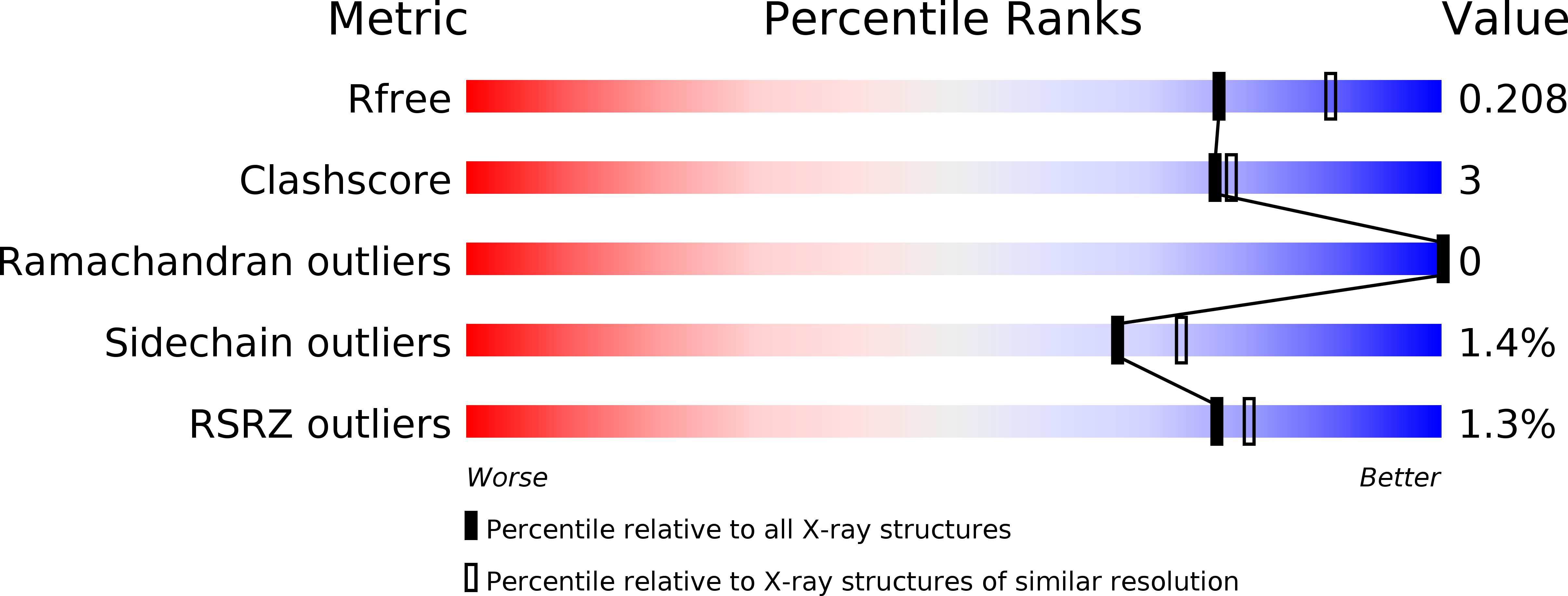
R-Value Free:
0.20
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
P 61 2 2