Deposition Date
2014-01-27
Release Date
2014-06-11
Last Version Date
2025-02-12
Entry Detail
PDB ID:
4OMF
Keywords:
Title:
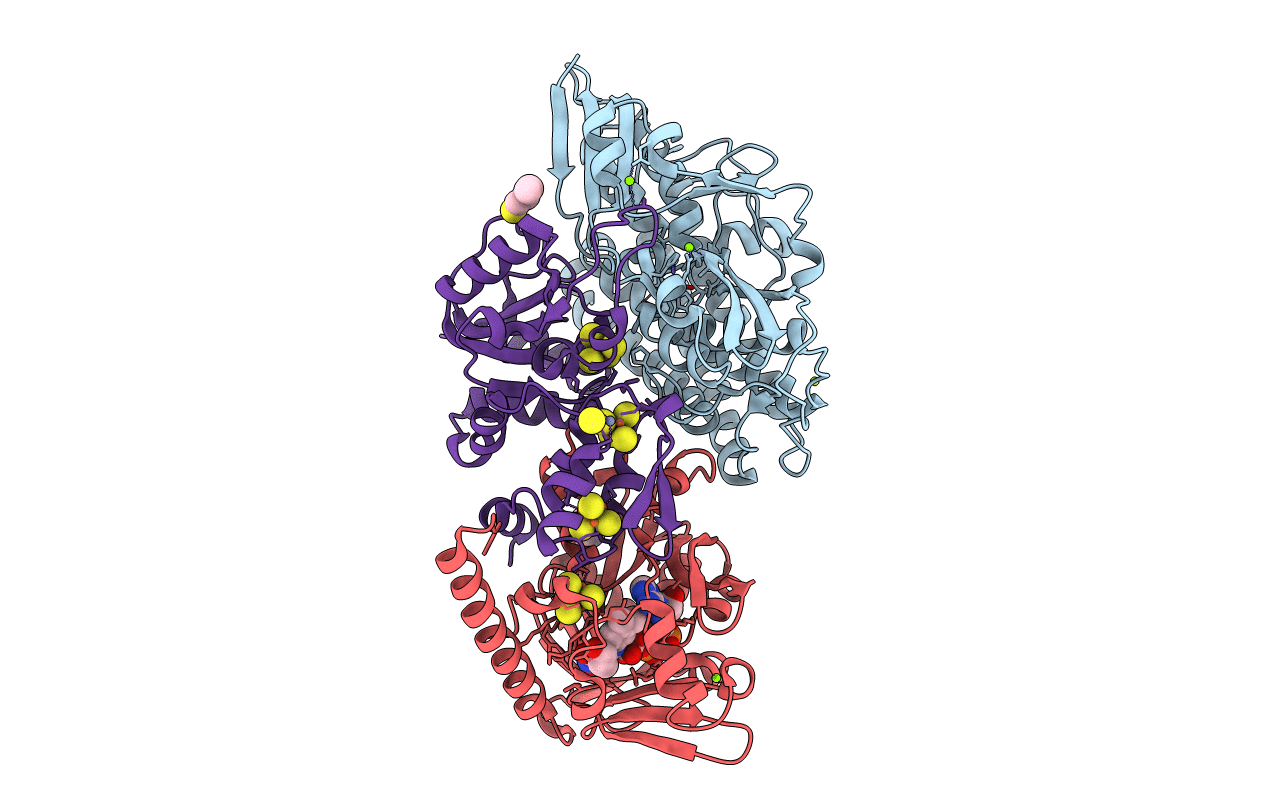
The F420-reducing [NiFe]-hydrogenase complex from Methanothermobacter marburgensis, the first X-ray structure of a group 3 family member
Biological Source:
Source Organism(s):
Methanothermobacter marburgensis (Taxon ID: 79929)
Method Details:
Experimental Method:
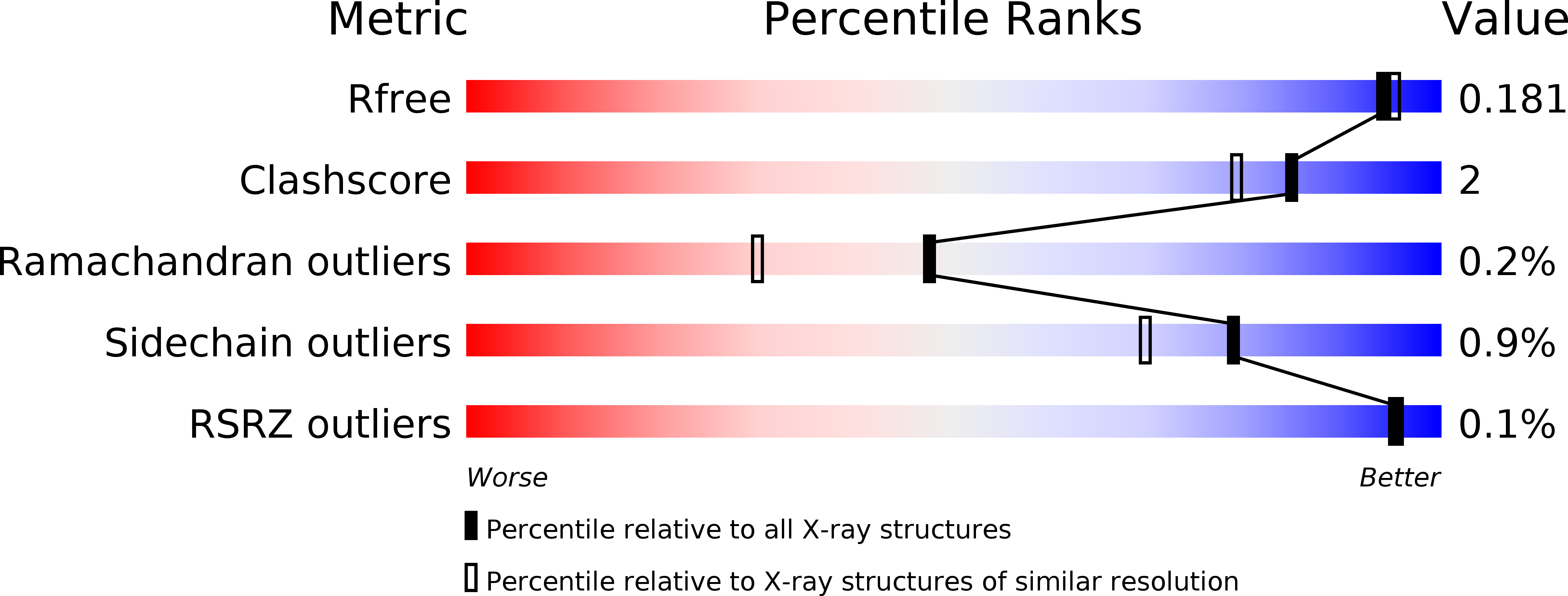
Resolution:
1.71 Å
R-Value Free:
0.18
R-Value Work:
0.15
R-Value Observed:
0.15
Space Group:
F 2 3