Deposition Date
2013-11-27
Release Date
2014-10-15
Last Version Date
2023-11-08
Entry Detail
Biological Source:
Source Organism:
Helicobacter pylori (Taxon ID: 85962)
Host Organism:
Method Details:
Experimental Method:
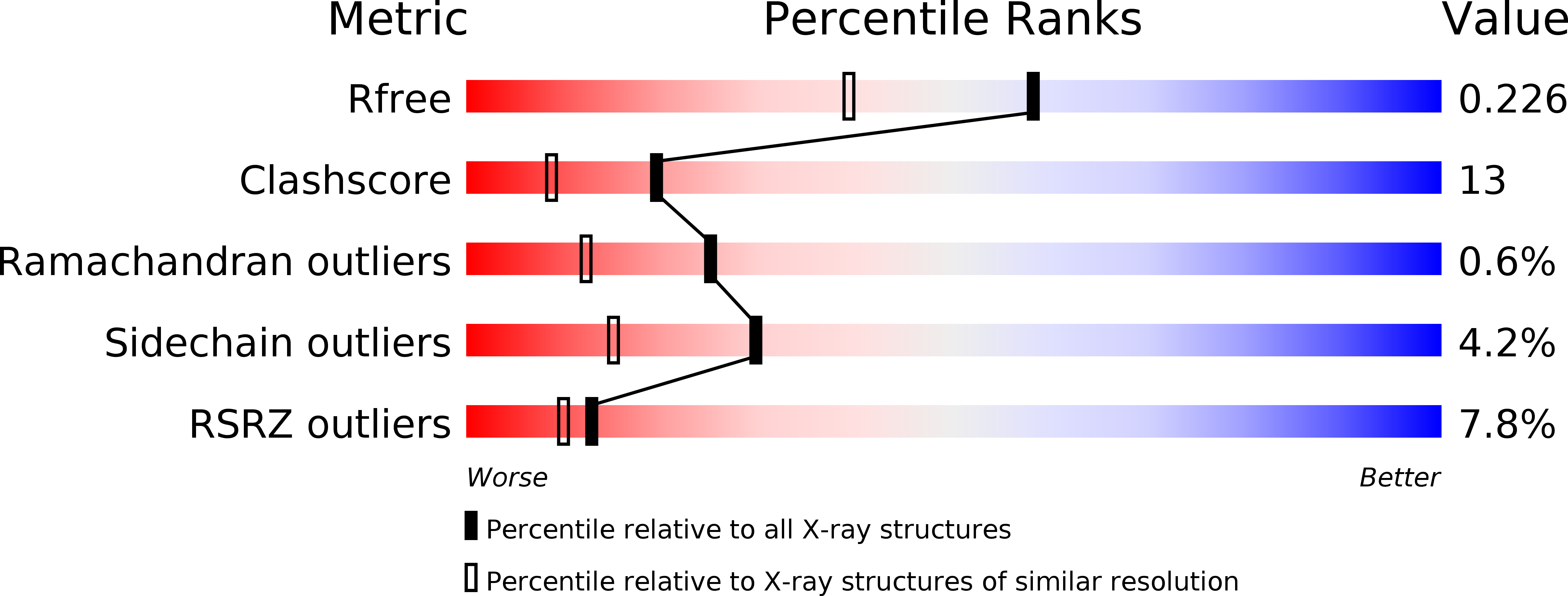
Resolution:
1.80 Å
R-Value Free:
0.22
R-Value Work:
0.18
R-Value Observed:
0.18
Space Group:
P 1 21 1