Deposition Date
2013-11-04
Release Date
2014-01-15
Last Version Date
2024-02-28
Entry Detail
PDB ID:
4NH7
Keywords:
Title:
Correlation between chemotype-dependent binding conformations of HSP90 alpha/beta and isoform selectivity
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
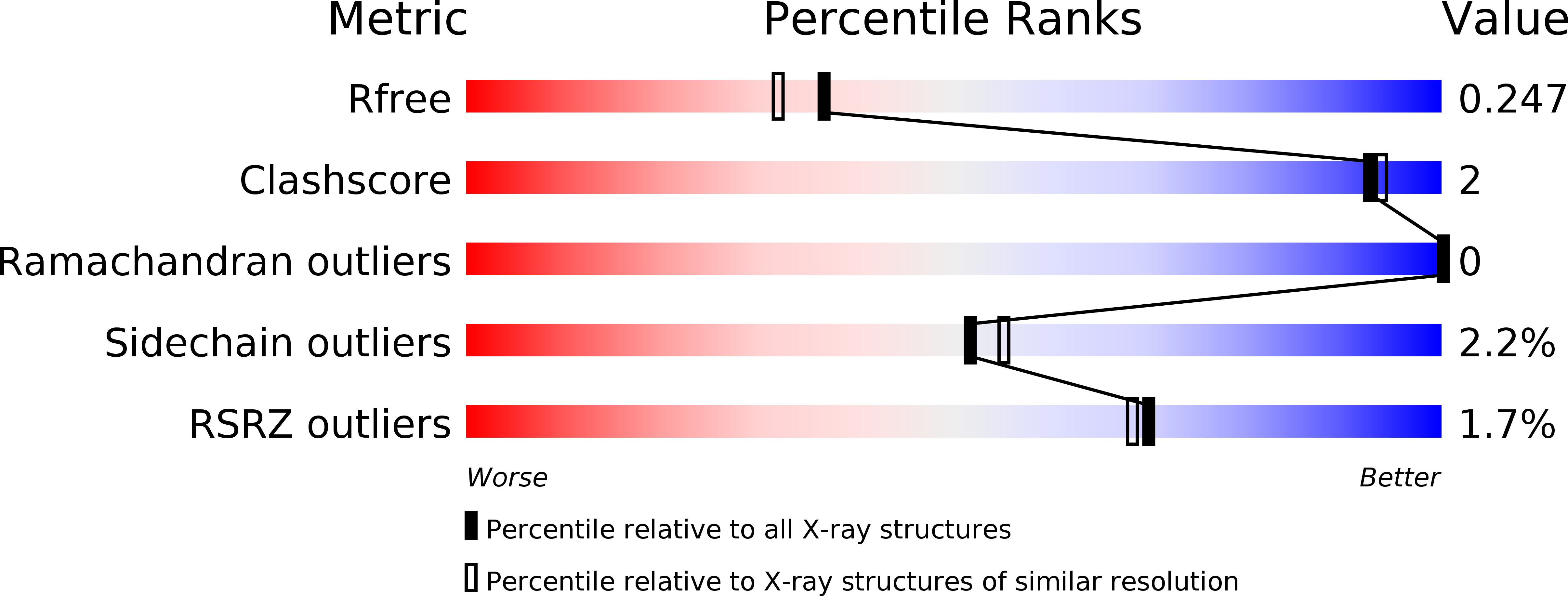
Resolution:
2.00 Å
R-Value Free:
0.24
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
C 1 2 1