Deposition Date
2013-09-04
Release Date
2013-11-06
Last Version Date
2023-09-20
Entry Detail
PDB ID:
4MJO
Keywords:
Title:
Human liver fructose-1,6-bisphosphatase(d-fructose-1,6-bisphosphate, 1-phosphohydrolase) (e.c.3.1.3.11) complexed with the allosteric inhibitor 3
Biological Source:
Source Organism:
Homo sapiens (Taxon ID: 9606)
Method Details:
Experimental Method:
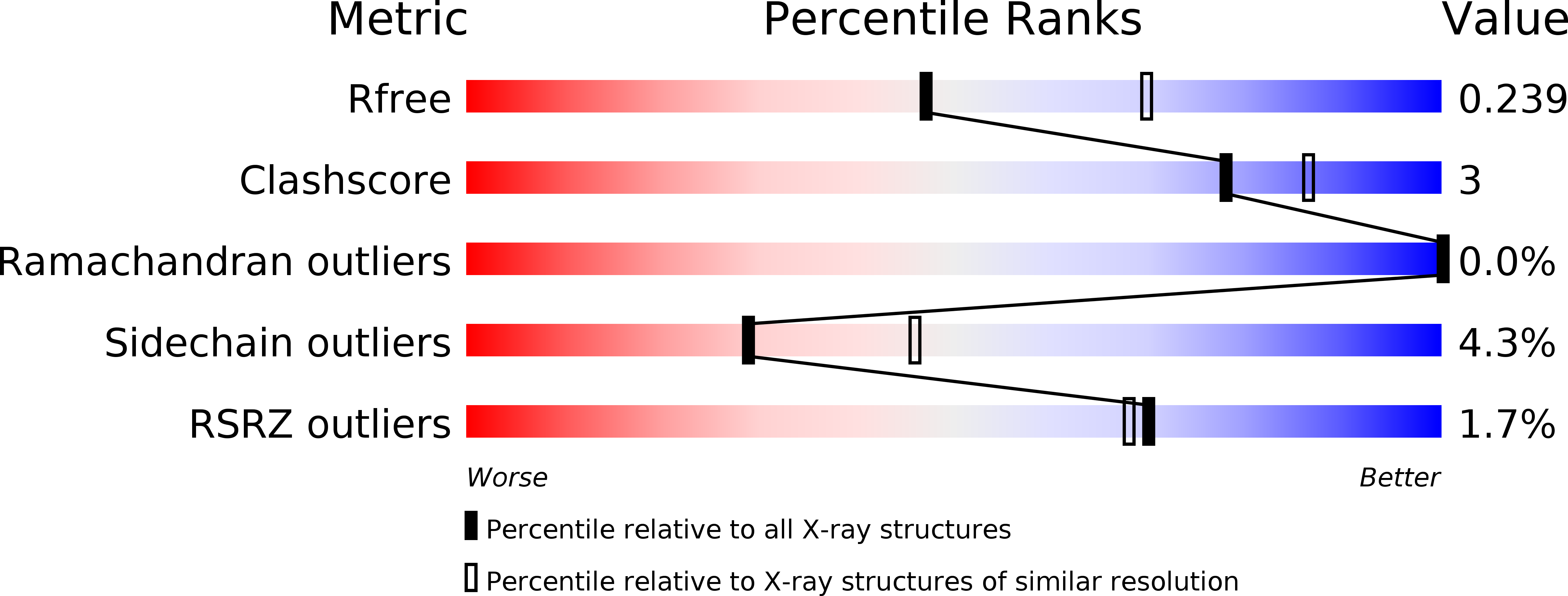
Resolution:
2.40 Å
R-Value Free:
0.23
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
P 1 21 1