Deposition Date
2013-05-02
Release Date
2014-05-14
Last Version Date
2024-02-28
Entry Detail
PDB ID:
4KIJ
Keywords:
Title:
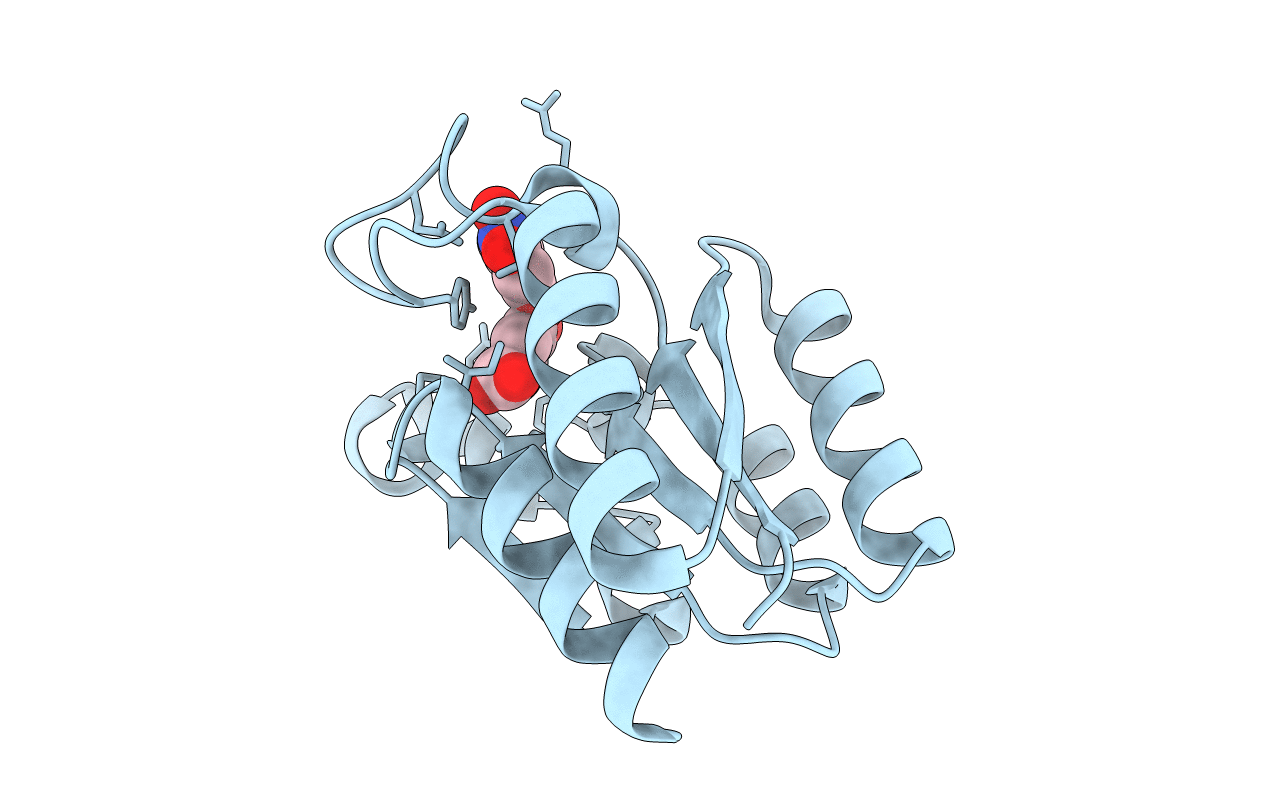
Design and structural analysis of aromatic inhibitors of type II dehydroquinase dehydratase from Mycobacterium tuberculosis - compound 35c [3,4-dihydroxy-5-(3-nitrophenoxy)benzoic acid]
Biological Source:
Source Organism(s):
Mycobacterium tuberculosis (Taxon ID: 1773)
Expression System(s):
Method Details:
Experimental Method:
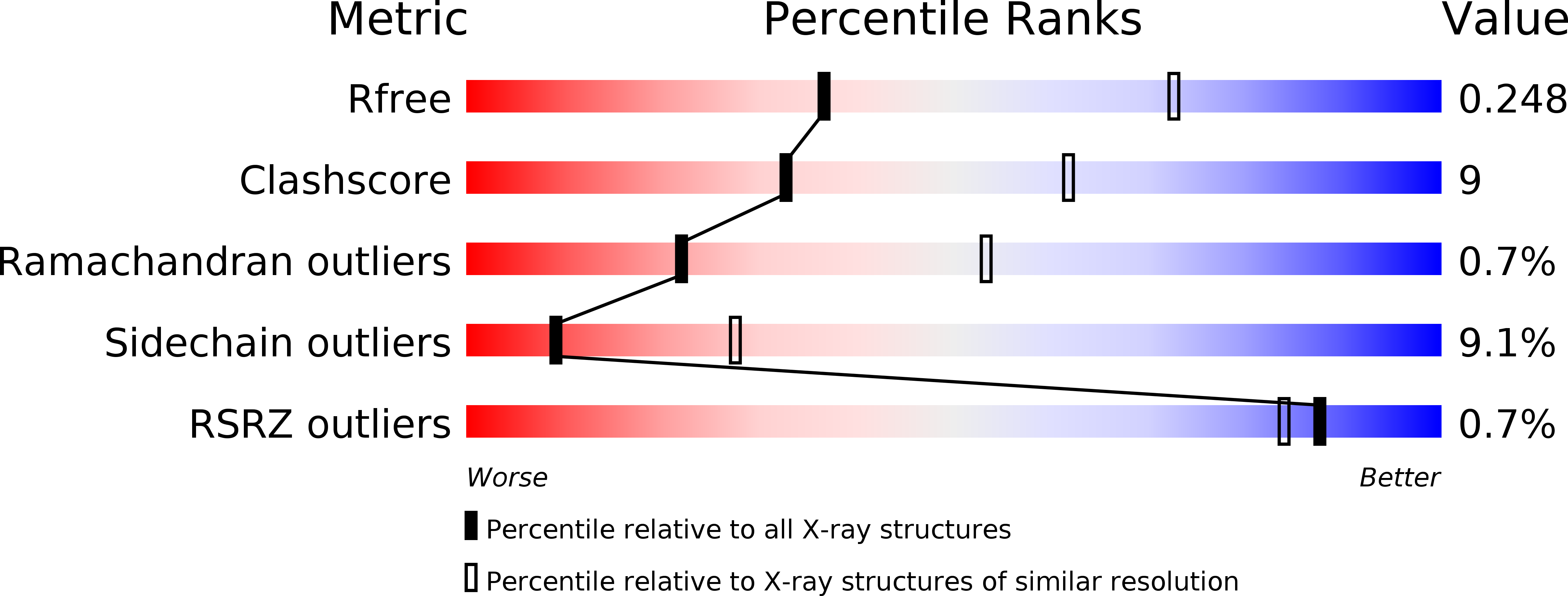
Resolution:
2.80 Å
R-Value Free:
0.24
R-Value Work:
0.19
R-Value Observed:
0.19
Space Group:
F 2 3