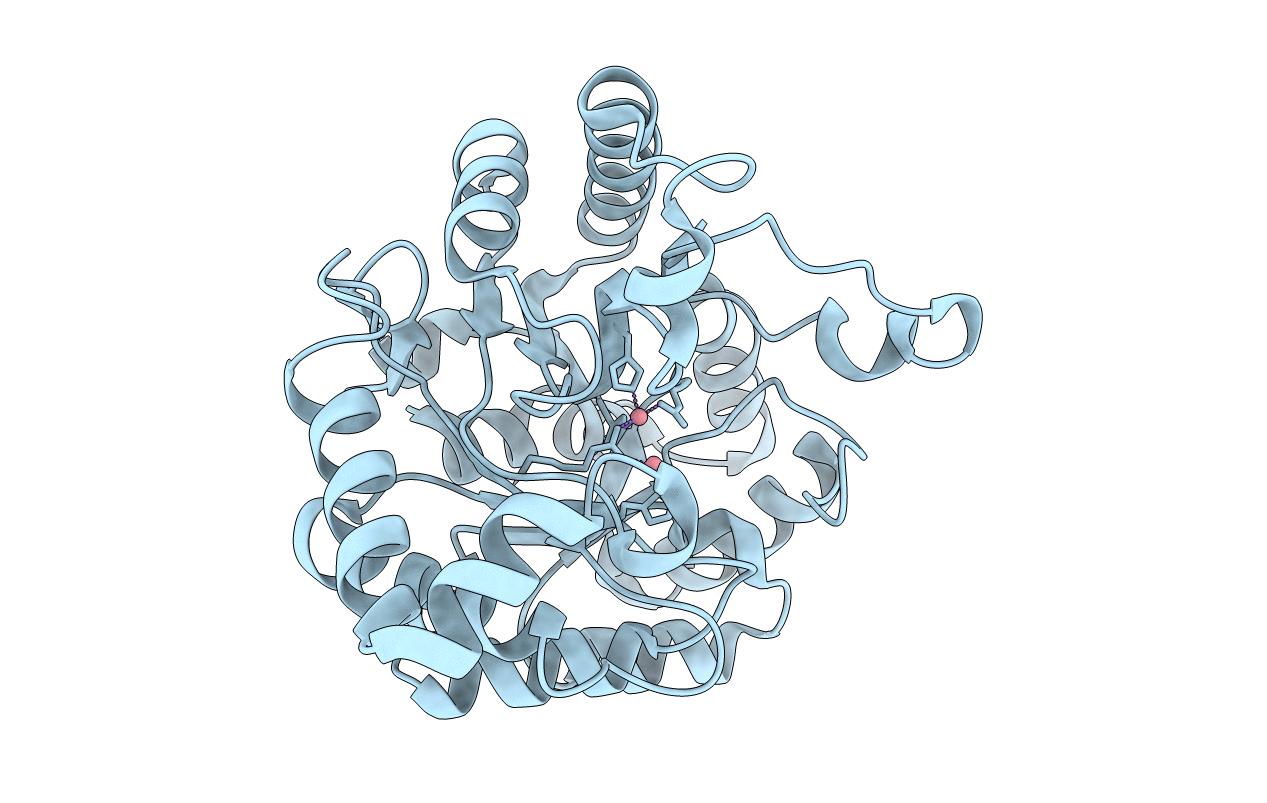
Deposition Date
2013-02-08
Release Date
2013-07-24
Last Version Date
2023-12-06
Entry Detail
PDB ID:
4J5N
Keywords:
Title:
Crystal Structure of a Deinococcus radiodurans PTE-like lactonase (drPLL) mutant Y28L/D71N/E101G/E179D/V235L/L270M
Biological Source:
Source Organism(s):
Deinococcus radiodurans (Taxon ID: 1299)
Expression System(s):
Method Details:
Experimental Method:
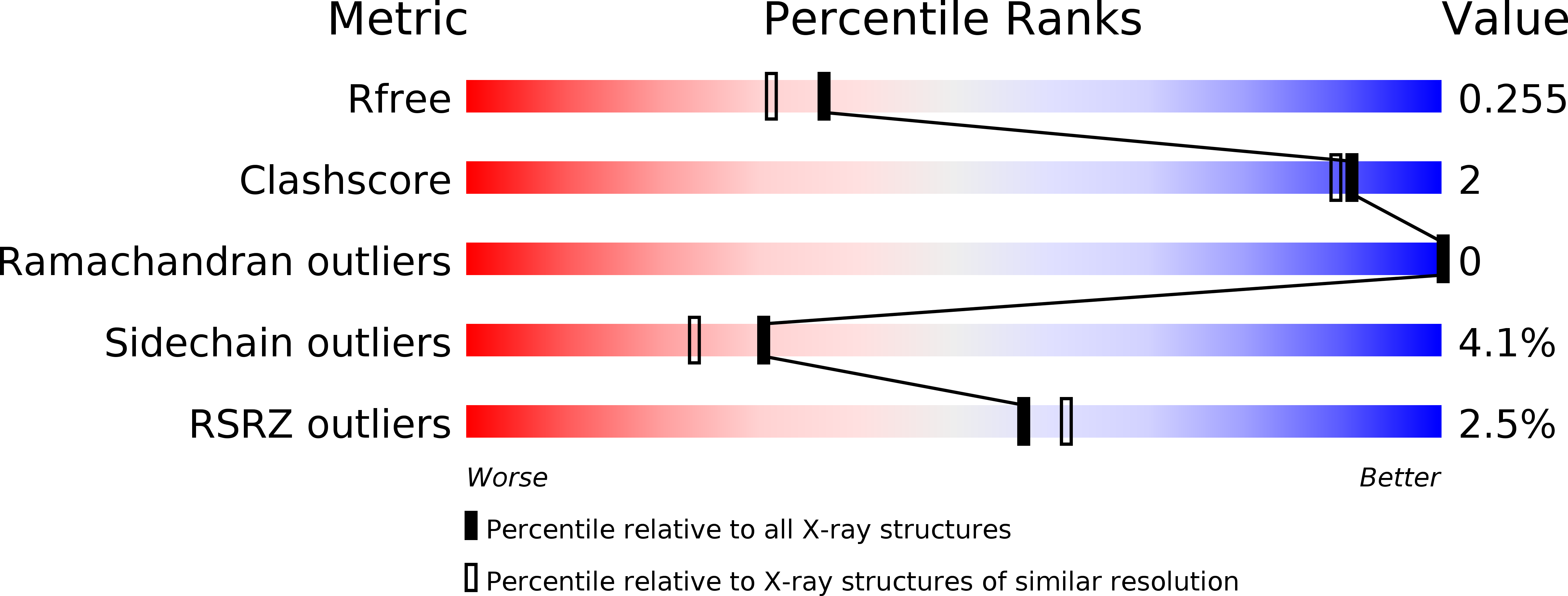
Resolution:
2.05 Å
R-Value Free:
0.26
R-Value Work:
0.23
Space Group:
P 31 2 1