Deposition Date
2013-01-21
Release Date
2013-03-06
Last Version Date
2024-11-20
Entry Detail
PDB ID:
4IUY
Keywords:
Title:
Crystal structure of short-chain dehydrogenase/reductase (apo-form) from A. baumannii clinical strain WM99C
Biological Source:
Source Organism:
Acinetobacter baumannii (Taxon ID: 405416)
Host Organism:
Method Details:
Experimental Method:
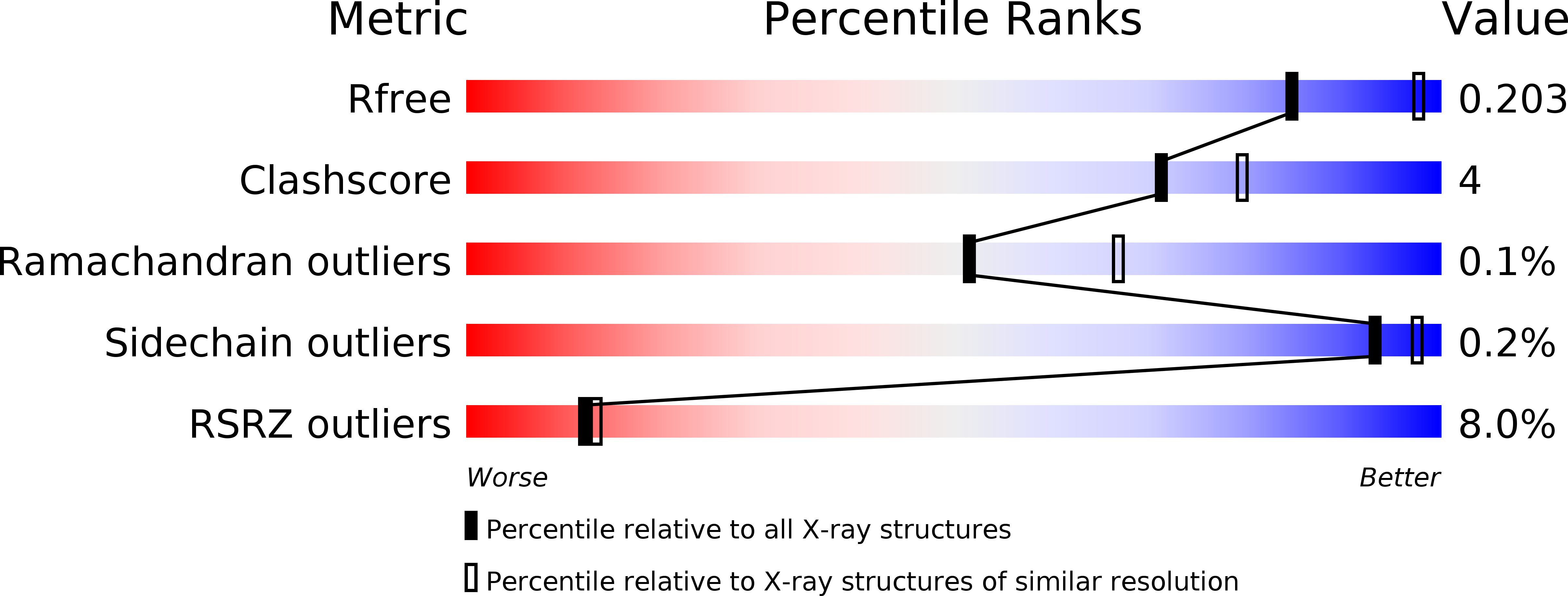
Resolution:
2.39 Å
R-Value Free:
0.20
R-Value Work:
0.15
R-Value Observed:
0.15
Space Group:
P 1 21 1