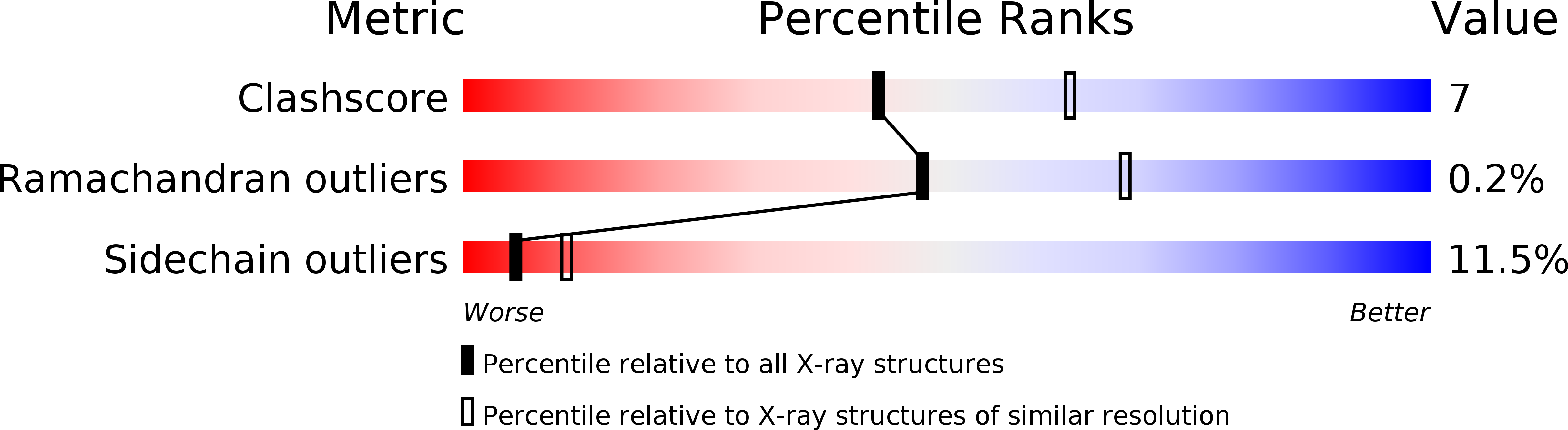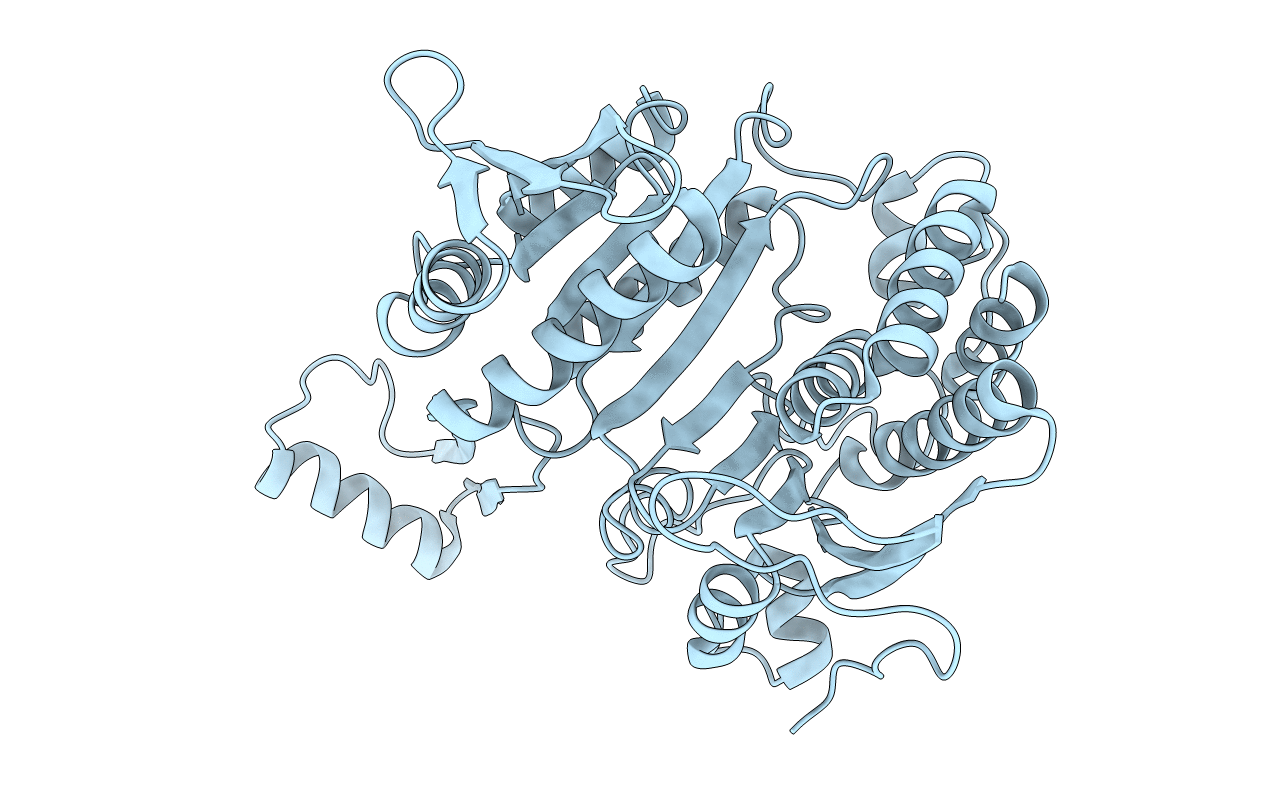
Deposition Date
1989-12-28
Release Date
1991-01-15
Last Version Date
2024-10-09
Entry Detail
PDB ID:
4ICD
Keywords:
Title:
REGULATION OF ISOCITRATE DEHYDROGENASE BY PHOSPHORYLATION INVOLVES NO LONG-RANGE CONFORMATIONAL CHANGE IN THE FREE ENZYME
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 562)
Method Details: