Deposition Date
2012-11-15
Release Date
2013-06-19
Last Version Date
2023-09-20
Entry Detail
PDB ID:
4HZM
Keywords:
Title:
Crystal structure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) bound to N-[(3S,4R,5R,6R)-4,5-dihydroxy-6-(hydroxymethyl)piperidin-3-yl]butanamide
Biological Source:
Source Organism(s):
Expression System(s):
Method Details:
Experimental Method:
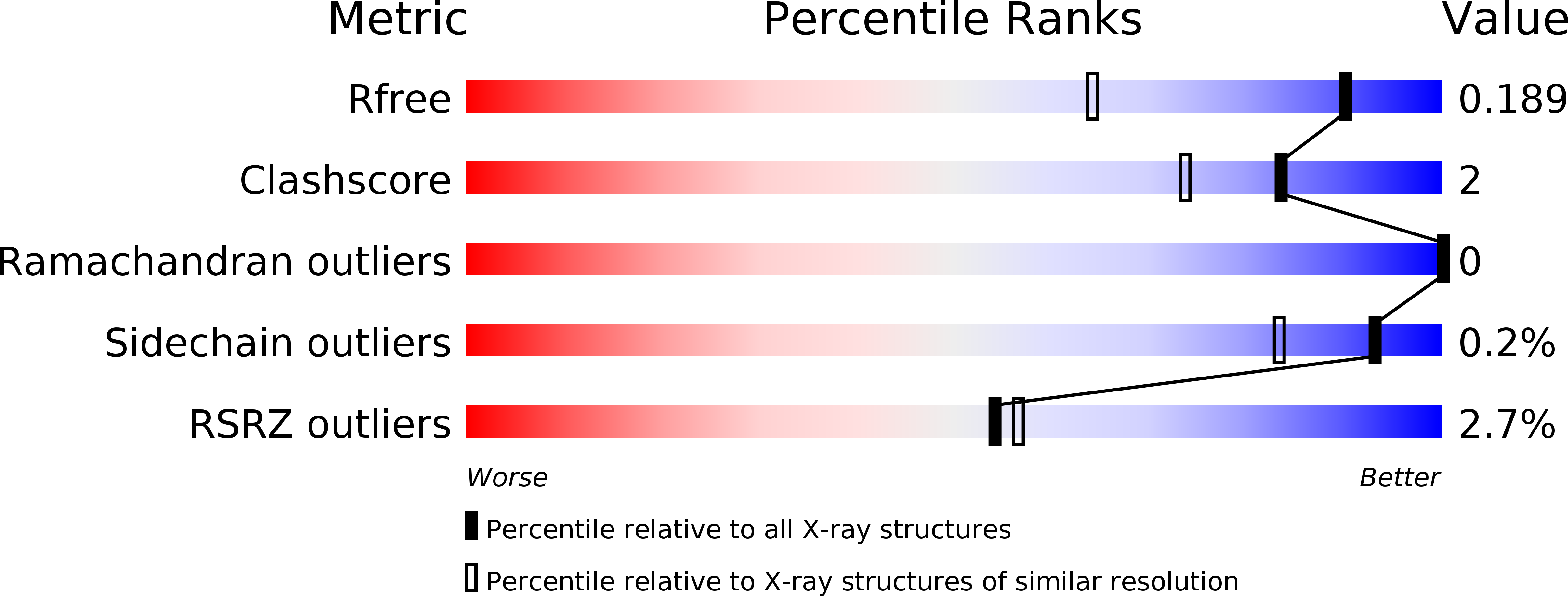
Resolution:
1.45 Å
R-Value Free:
0.19
R-Value Work:
0.16
R-Value Observed:
0.17
Space Group:
P 1 21 1