Deposition Date
2012-11-14
Release Date
2013-09-11
Last Version Date
2024-02-28
Entry Detail
PDB ID:
4HYV
Keywords:
Title:
Pyruvate kinase (PYK) from Trypanosoma brucei in the presence of Magnesium, PEP and F26BP
Biological Source:
Source Organism:
Trypanosoma brucei brucei (Taxon ID: 5702)
Host Organism:
Method Details:
Experimental Method:
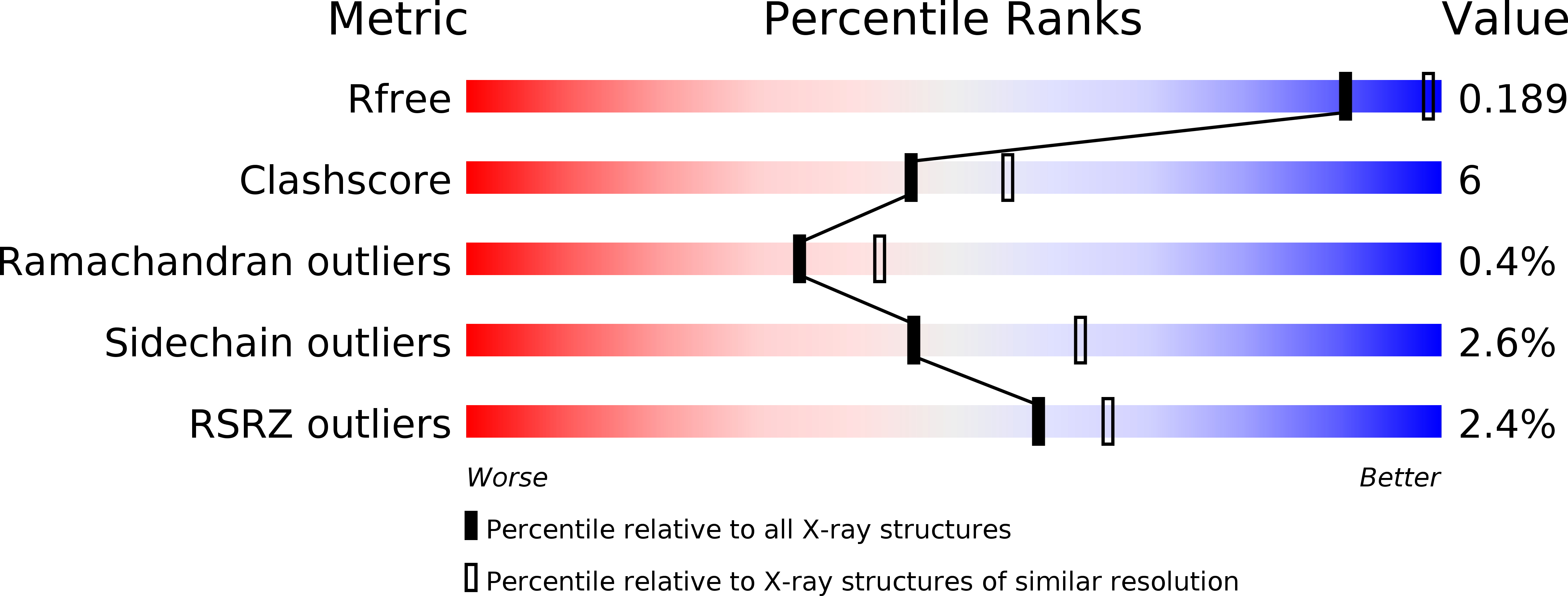
Resolution:
2.30 Å
R-Value Free:
0.18
R-Value Work:
0.14
R-Value Observed:
0.14
Space Group:
I 2 2 2