Deposition Date
2012-08-11
Release Date
2013-06-05
Last Version Date
2024-02-28
Entry Detail
PDB ID:
4GKK
Keywords:
Title:
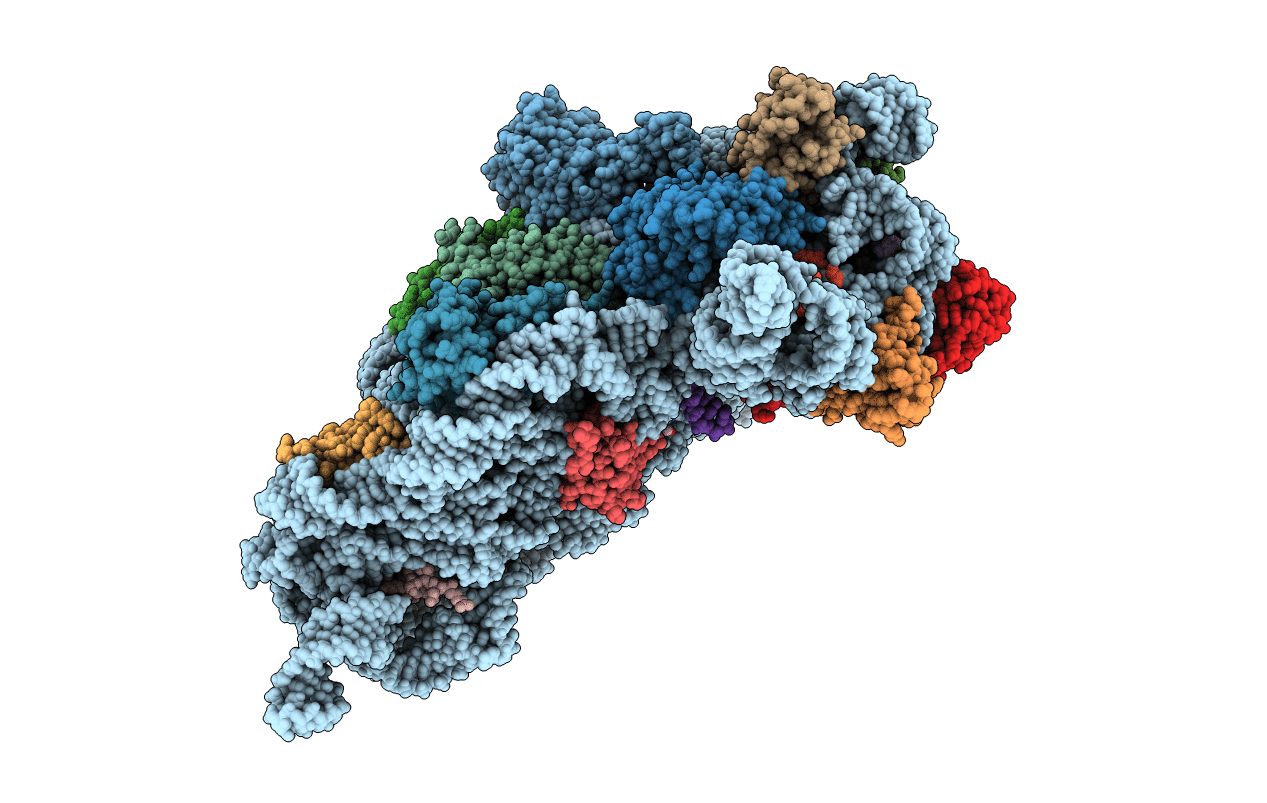
Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a human mitochondrial anticodon stem loop (ASL) of transfer RNA Methionine (TRNAMET) bound to an mRNA with an AUA-codon in the A-site and paromomycin
Biological Source:
Source Organism(s):
Synthetic (Taxon ID: 32630)
Thermus thermophilus (Taxon ID: 300852)
Thermus thermophilus (Taxon ID: 300852)
Method Details:
Experimental Method:
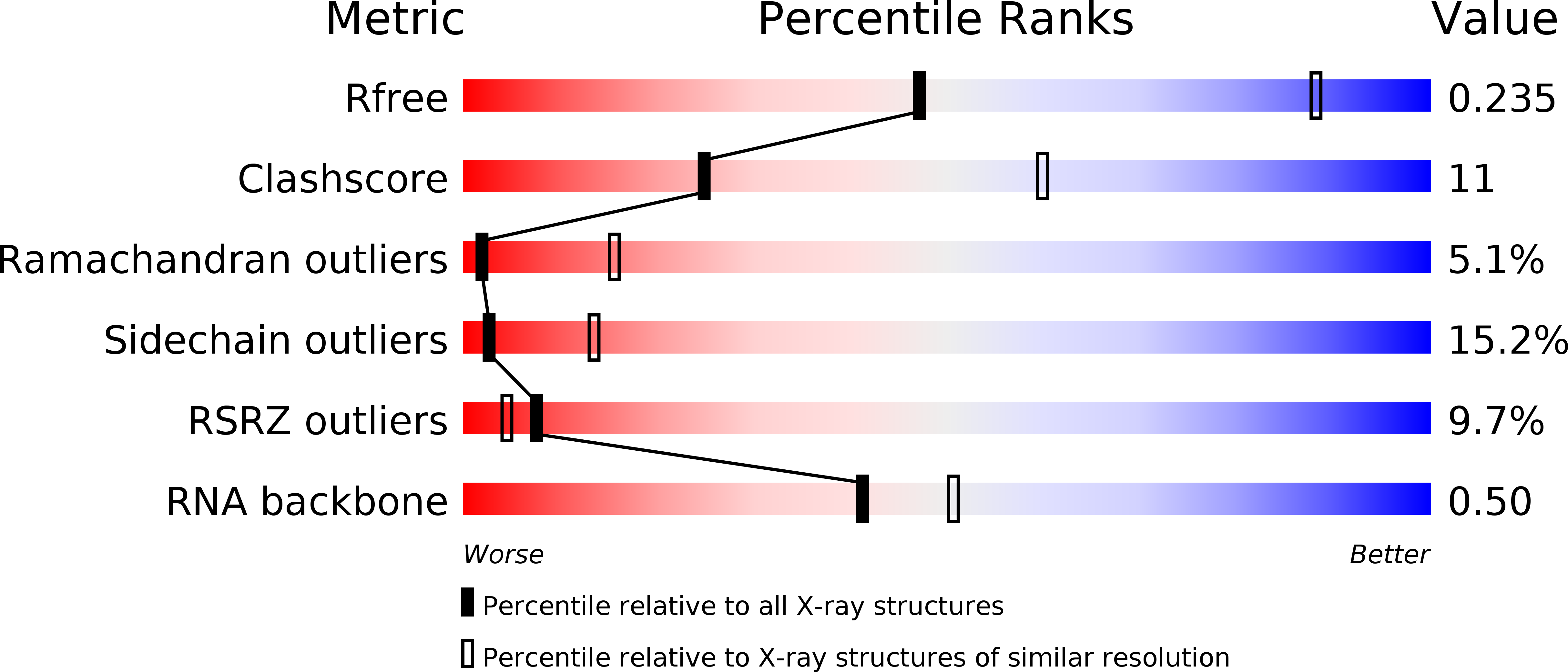
Resolution:
3.20 Å
R-Value Free:
0.23
R-Value Work:
0.19
R-Value Observed:
0.19
Space Group:
P 41 21 2