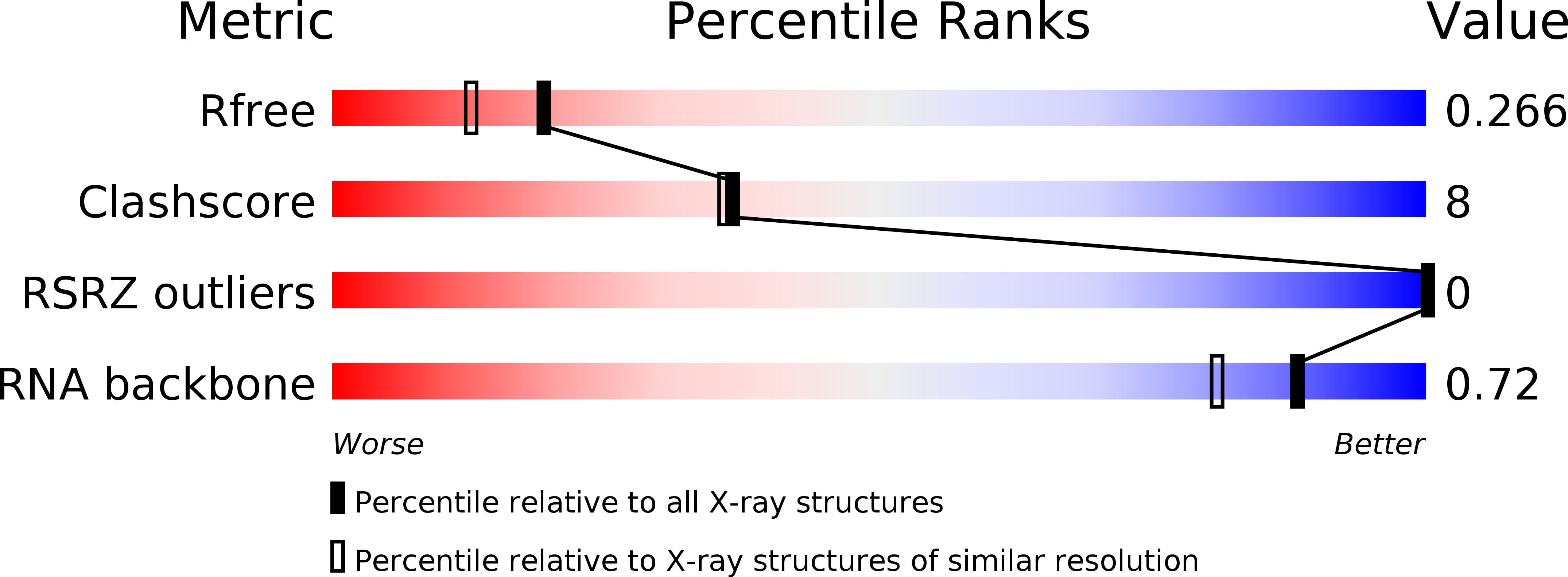
Deposition Date
2012-05-18
Release Date
2012-08-15
Last Version Date
2024-03-20
Entry Detail
PDB ID:
4F8U
Keywords:
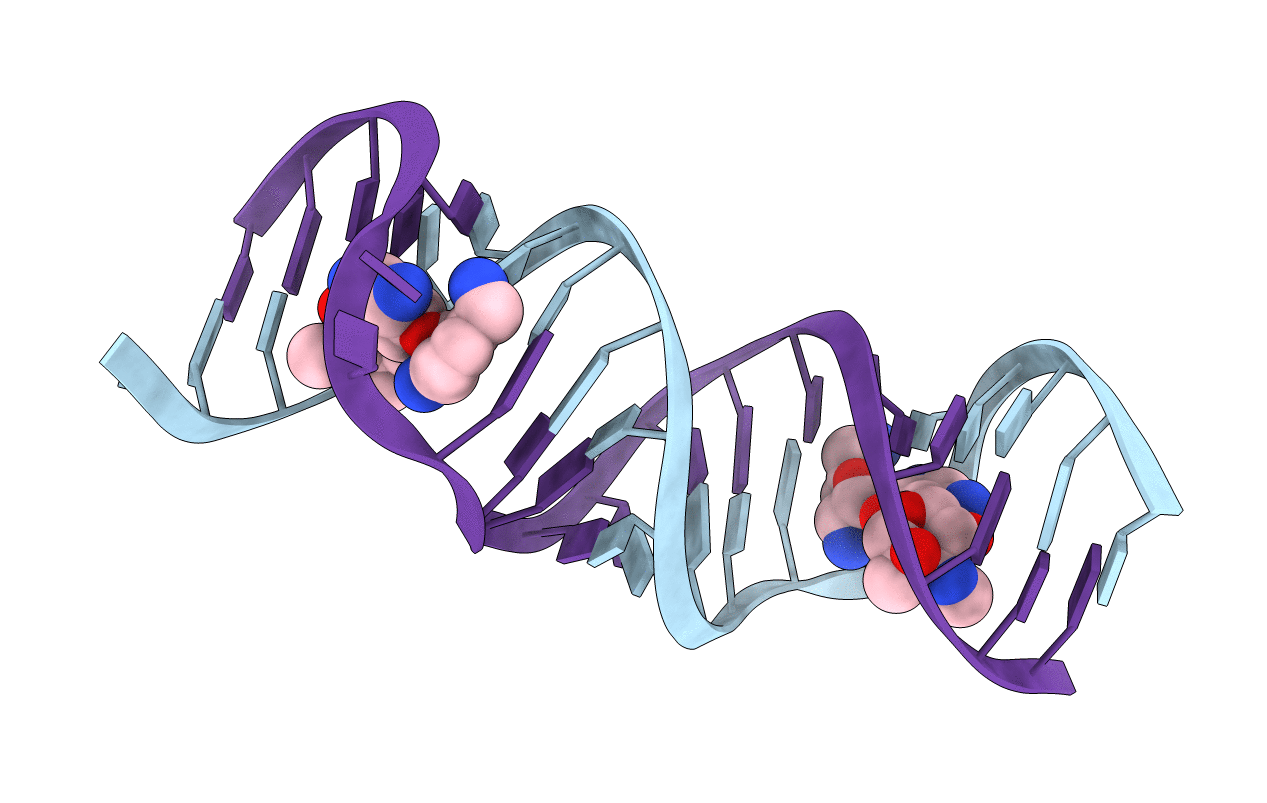
Title:
Crystal structure of the bacterial ribosomal decoding site in complex with sisomicin (C2 form)
Method Details:
Experimental Method:
Resolution:
2.00 Å
R-Value Free:
0.26
R-Value Work:
0.25
Space Group:
C 1 2 1