Deposition Date
2014-11-06
Release Date
2015-02-04
Last Version Date
2024-05-08
Entry Detail
PDB ID:
4D5N
Keywords:
Title:
Cryo-EM structures of ribosomal 80S complexes with termination factors and cricket paralysis virus IRES reveal the IRES in the translocated state
Biological Source:
Source Organism(s):
HOMO SAPIENS (Taxon ID: 9606)
CRICKET PARALYSIS VIRUS (Taxon ID: 12136)
CRICKET PARALYSIS VIRUS (Taxon ID: 12136)
Expression System(s):
Method Details:
Experimental Method:
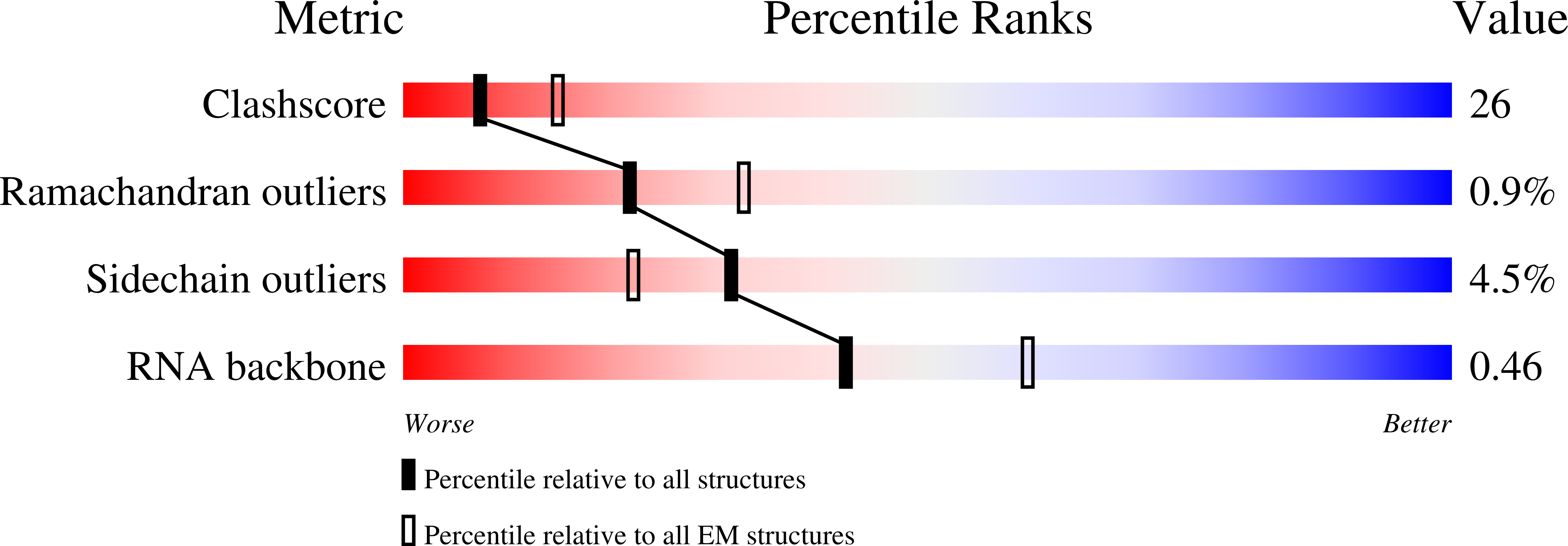
Resolution:
9.00 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE