Deposition Date
2014-04-17
Release Date
2014-06-11
Last Version Date
2024-05-08
Entry Detail
PDB ID:
4CZD
Keywords:
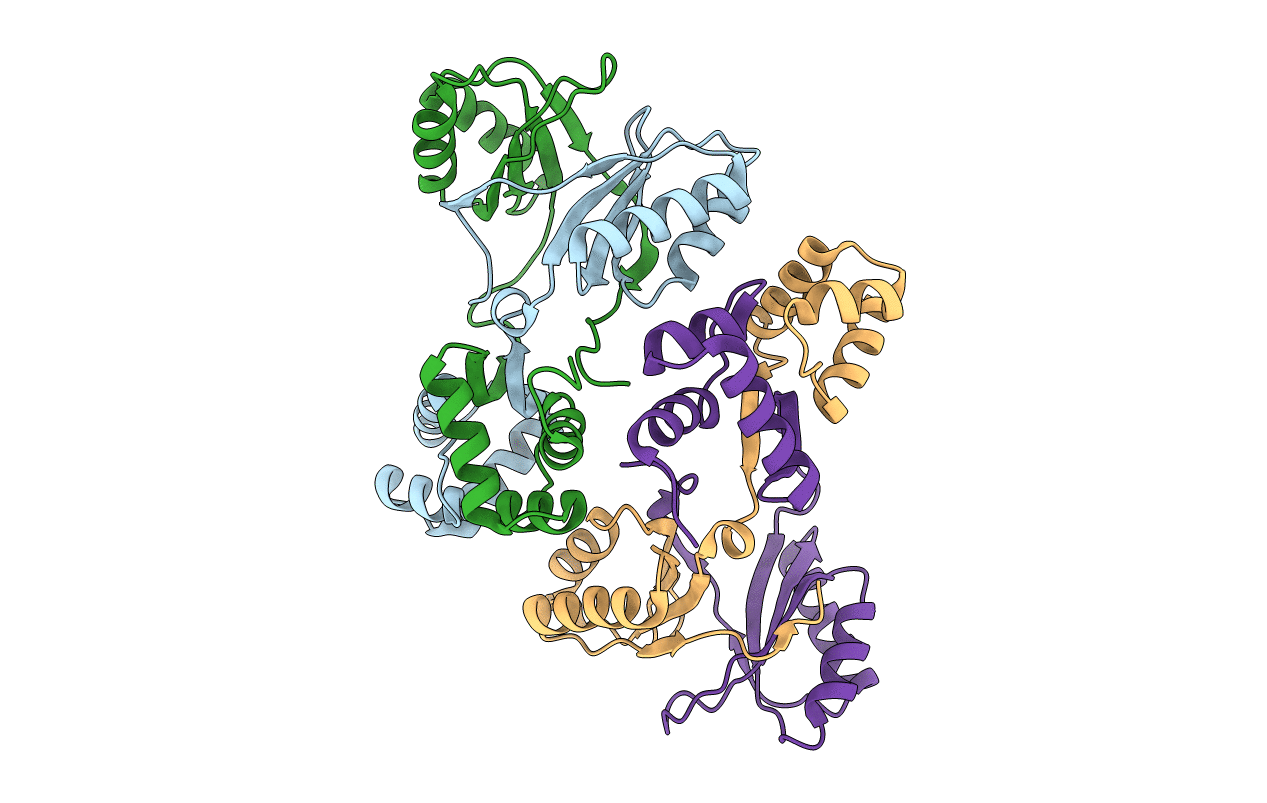
Title:
Sirohaem decarboxylase AhbA/B - an enzyme with structural homology to the Lrp/AsnC transcription factor family that is part of the alternative haem biosynthesis pathway.
Biological Source:
Source Organism(s):
DESULFOVIBRIO DESULFURICANS (Taxon ID: 876)
Expression System(s):
Method Details:
Experimental Method:
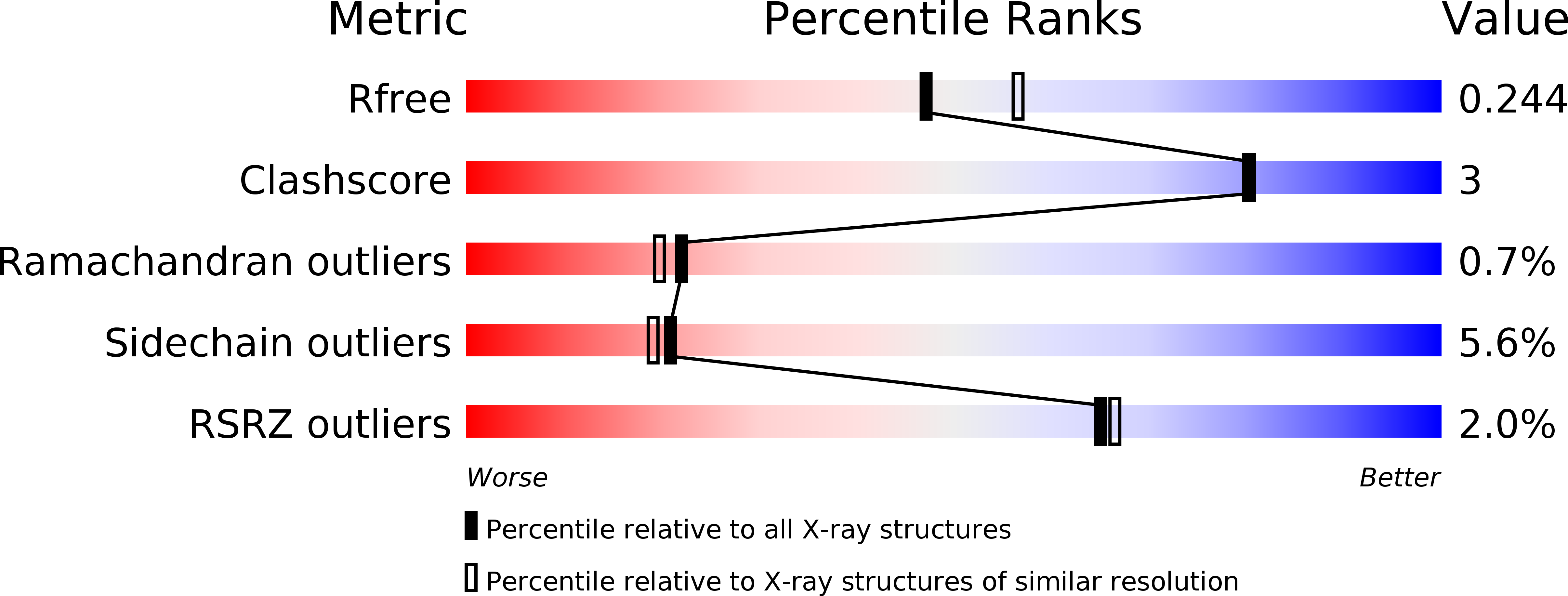
Resolution:
2.23 Å
R-Value Free:
0.23
R-Value Work:
0.19
R-Value Observed:
0.19
Space Group:
P 21 21 21