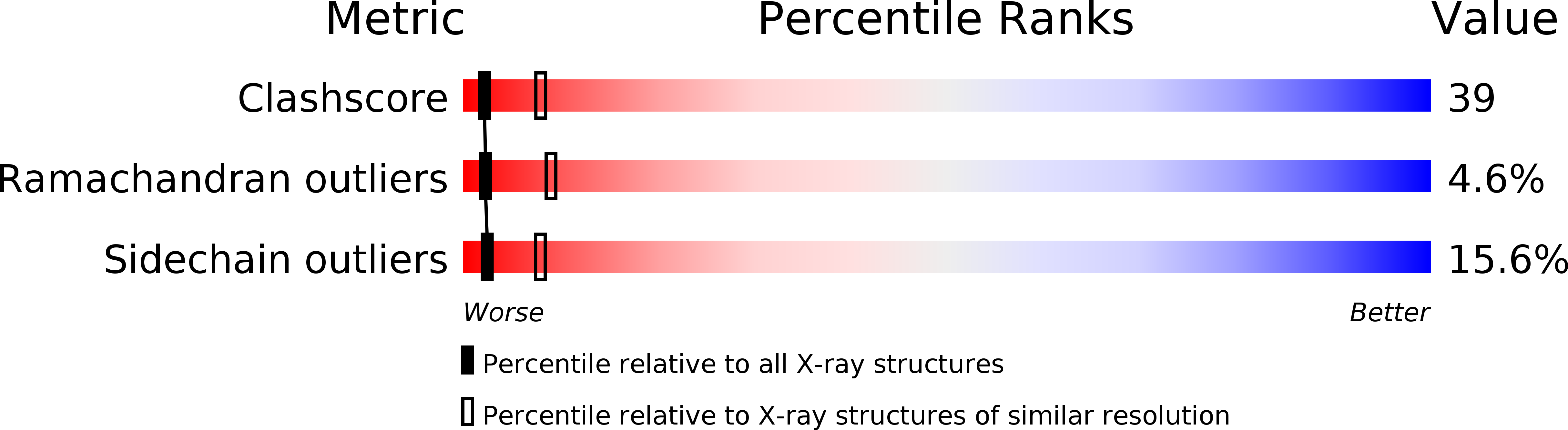
Deposition Date
1984-01-27
Release Date
1984-07-20
Last Version Date
2024-02-28
Entry Detail
PDB ID:
4CTS
Keywords:
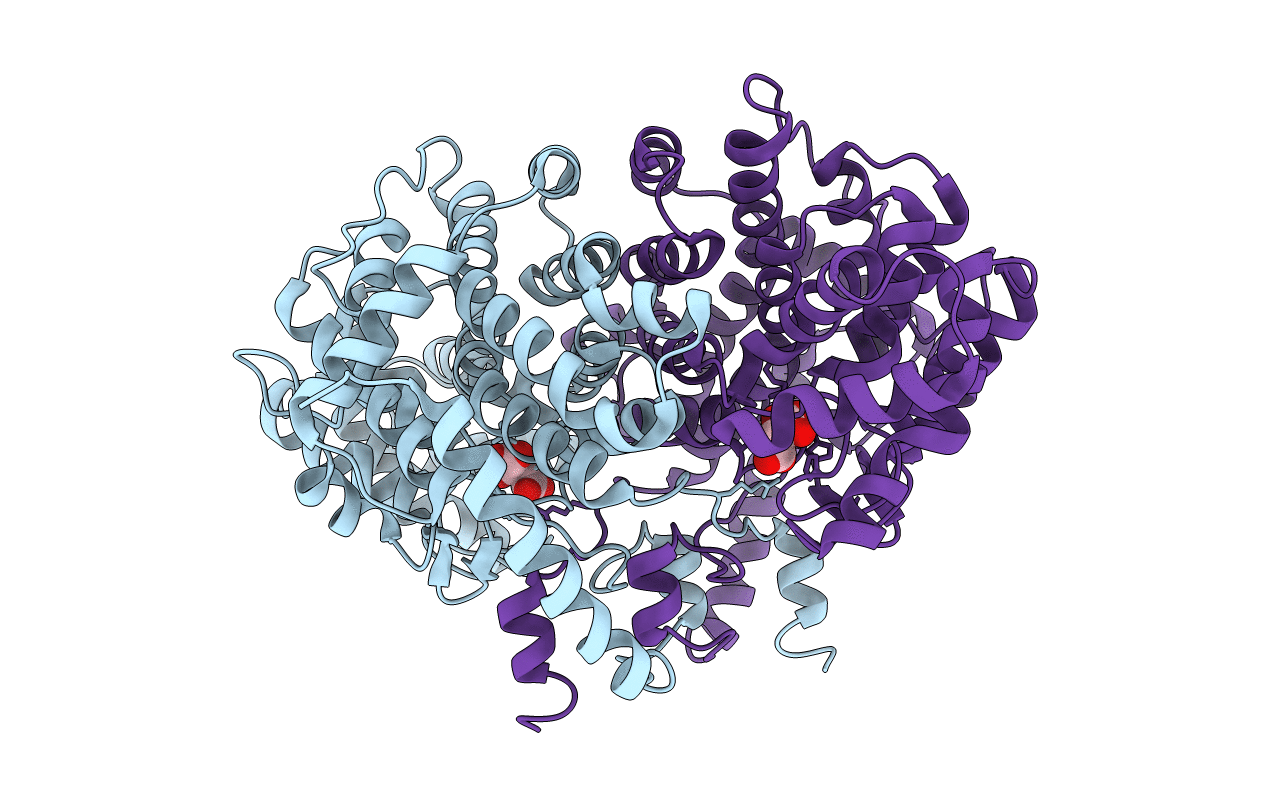
Title:
CRYSTAL STRUCTURE ANALYSIS AND MOLECULAR MODEL OF A COMPLEX OF CITRATE SYNTHASE WITH OXALOACETATE AND S-ACETONYL-COENZYME A
Biological Source:
Source Organism(s):
Sus scrofa (Taxon ID: 9823)
Method Details: