Deposition Date
2013-12-21
Release Date
2015-02-11
Last Version Date
2023-12-20
Entry Detail
PDB ID:
4CJN
Keywords:
Title:
Crystal structure of PBP2a from MRSA in complex with quinazolinone ligand
Biological Source:
Source Organism:
STAPHYLOCOCCUS AUREUS SUBSP. AUREUS MU50 (Taxon ID: 158878)
Host Organism:
Method Details:
Experimental Method:
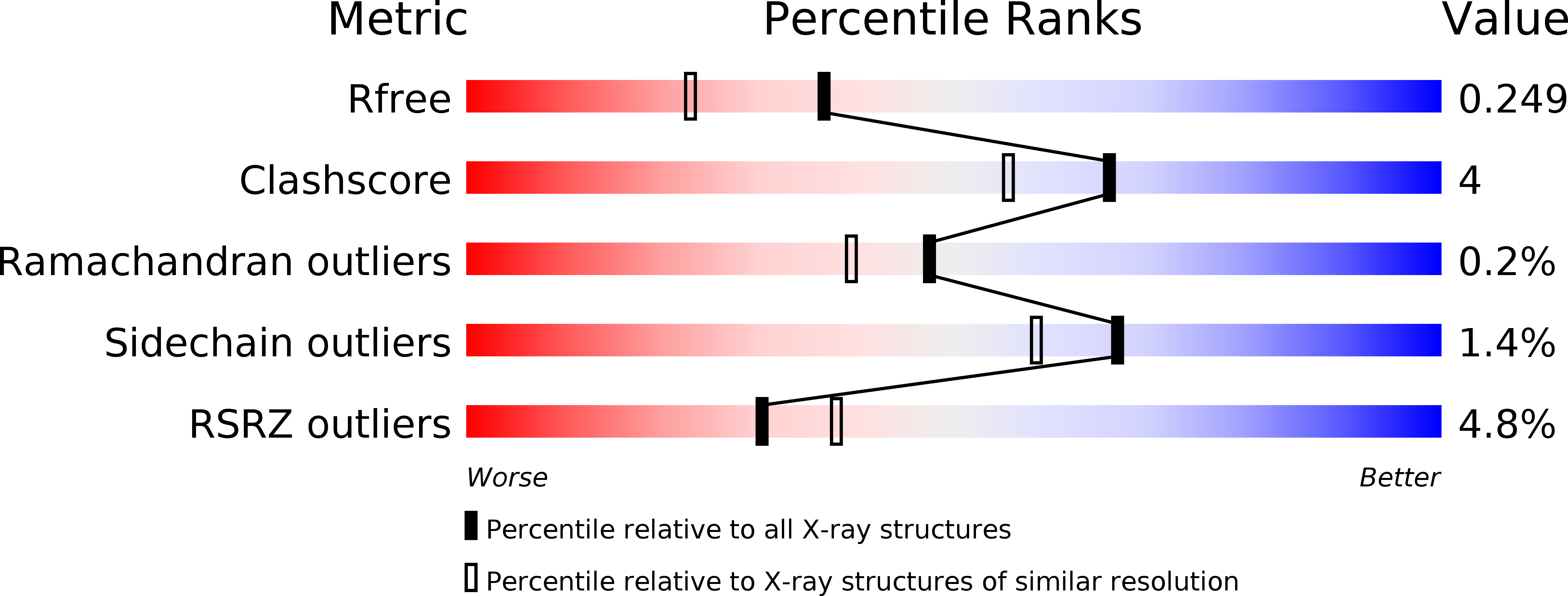
Resolution:
1.95 Å
R-Value Free:
0.24
R-Value Work:
0.19
R-Value Observed:
0.20
Space Group:
P 21 21 21