Deposition Date
2013-04-09
Release Date
2013-05-29
Last Version Date
2024-05-01
Entry Detail
PDB ID:
4BI6
Keywords:
Title:
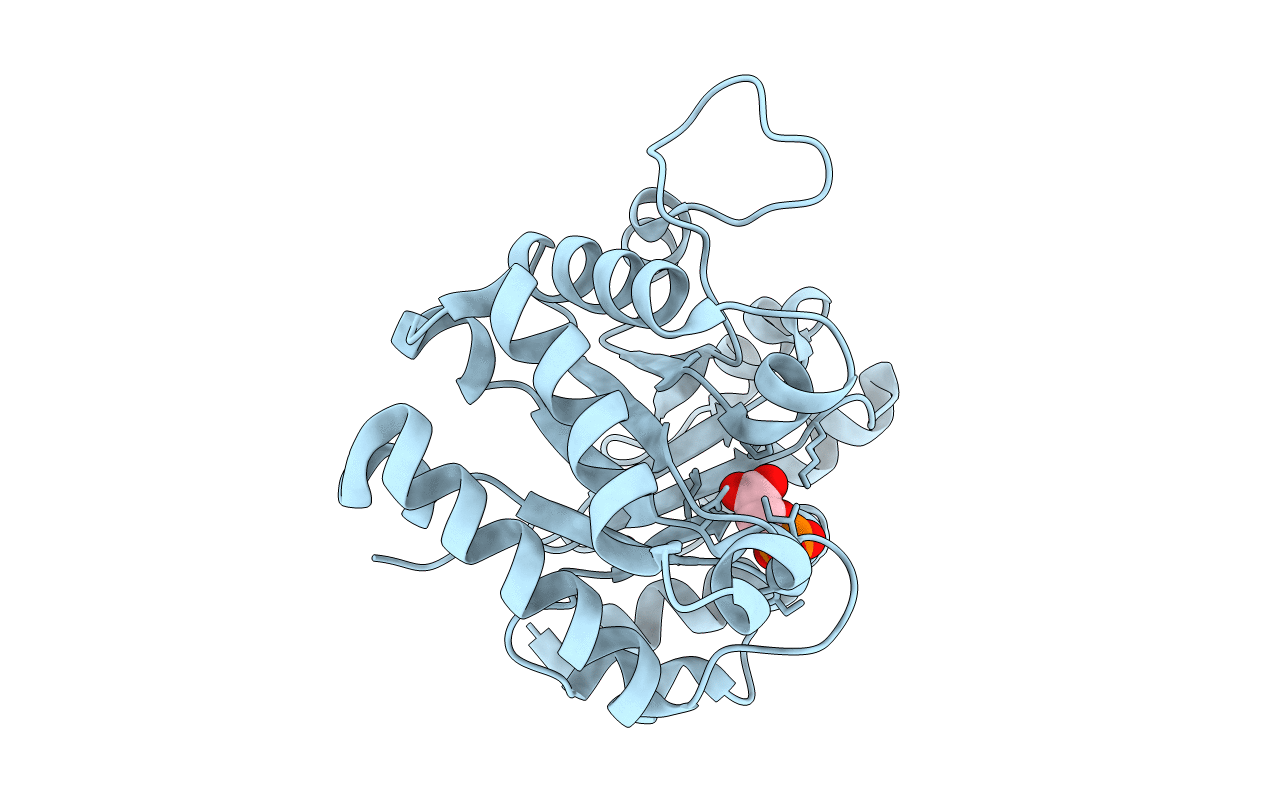
CRYSTAL STRUCTURE OF A TRIPLE MUTANT (A198V, C202A AND C222N) OF TRIOSEPHOSPHATE ISOMERASE FROM GIARDIA LAMBLIA. COMPLEXED WITH 2- PHOSPHOGLYCOLIC ACID
Biological Source:
Source Organism(s):
GIARDIA INTESTINALIS (Taxon ID: 5741)
Expression System(s):
Method Details:
Experimental Method:
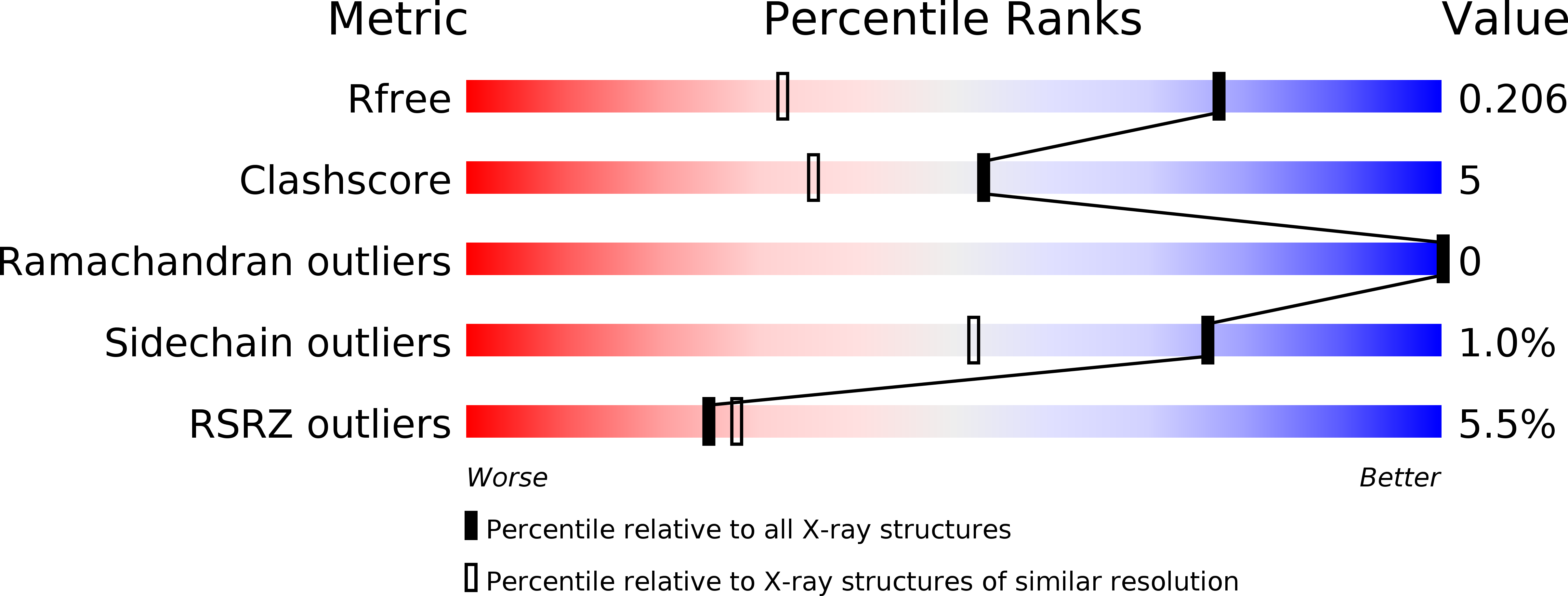
Resolution:
1.45 Å
R-Value Free:
0.20
R-Value Work:
0.18
R-Value Observed:
0.19
Space Group:
I 2 2 2