Deposition Date
2012-08-07
Release Date
2012-10-24
Last Version Date
2023-12-20
Entry Detail
PDB ID:
4B5P
Keywords:
Title:
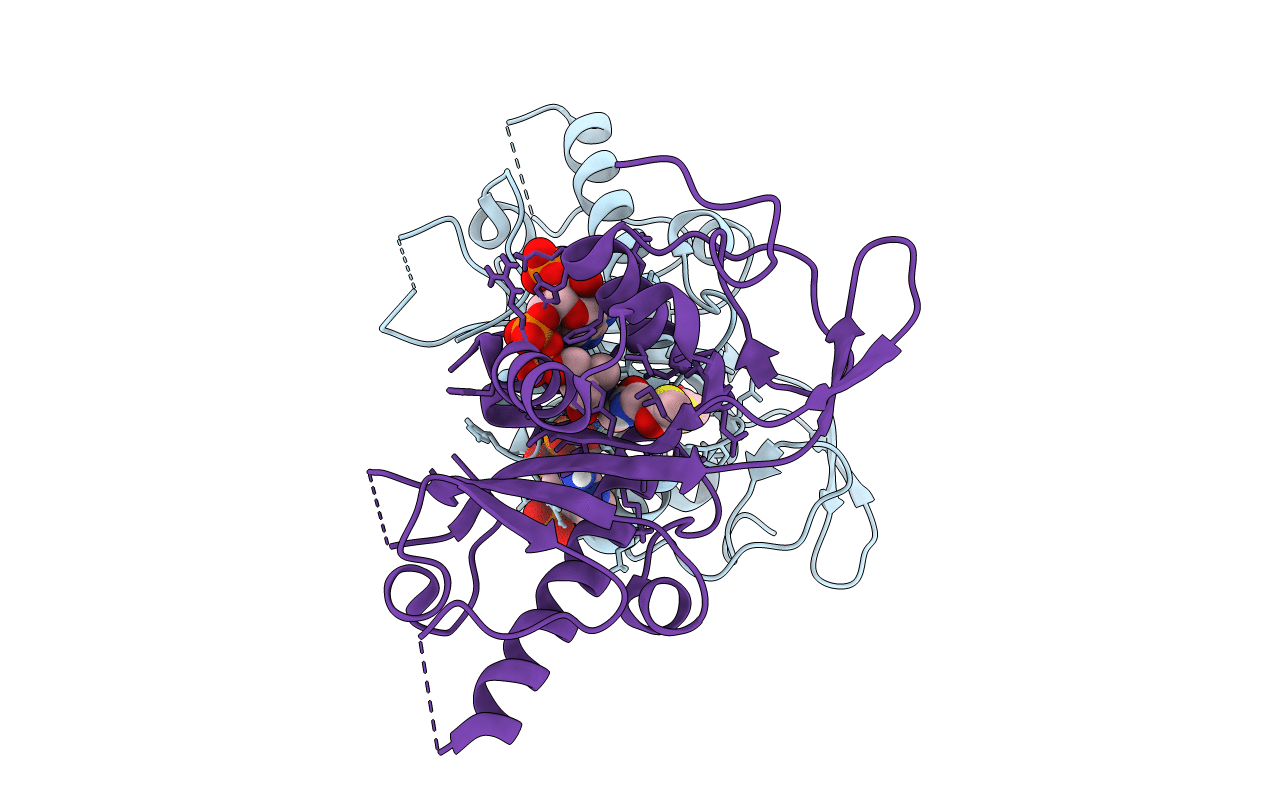
Crystal structure of human alpha tubulin acetyltransferase catalytic domain Q58A variant
Biological Source:
Source Organism(s):
HOMO SAPIENS (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
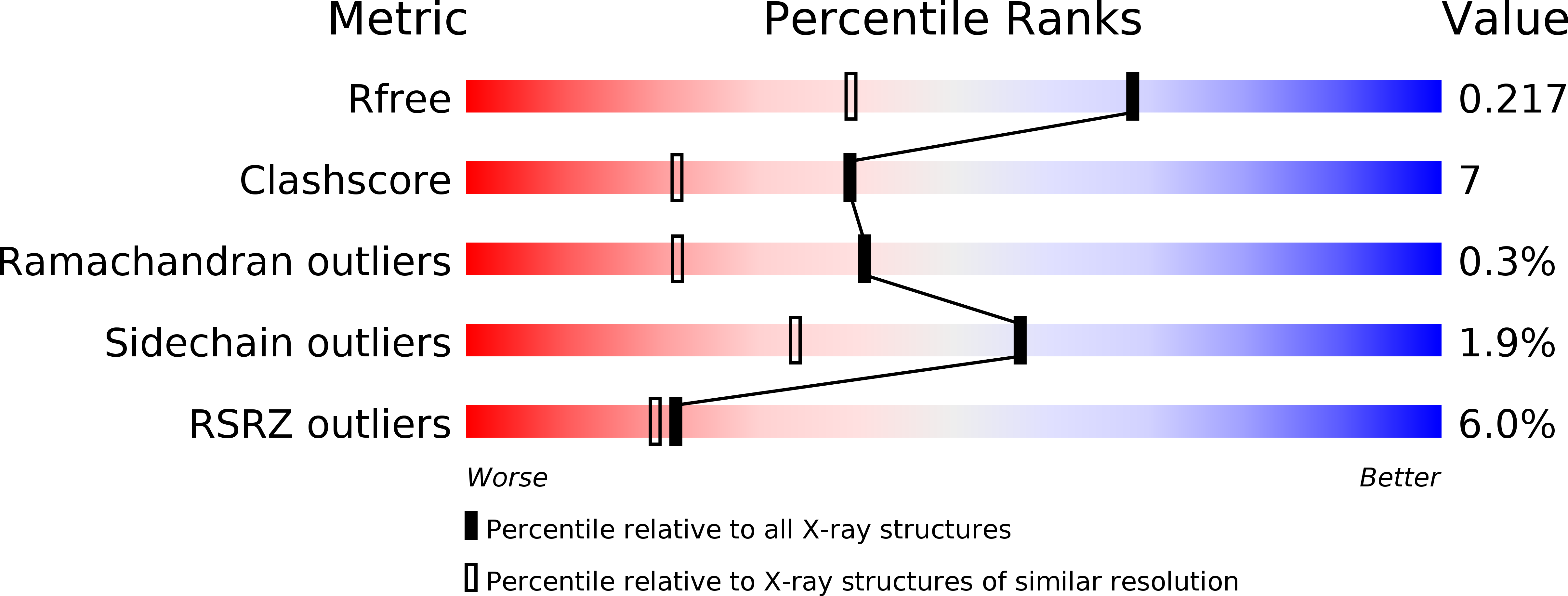
Resolution:
1.60 Å
R-Value Free:
0.22
R-Value Work:
0.18
R-Value Observed:
0.18
Space Group:
P 21 21 21