Deposition Date
2012-05-03
Release Date
2012-06-27
Last Version Date
2023-12-20
Entry Detail
PDB ID:
4ASU
Keywords:
Title:
F1-ATPase in which all three catalytic sites contain bound nucleotide, with magnesium ion released in the Empty site
Biological Source:
Source Organism(s):
BOS TAURUS (Taxon ID: 9913)
Method Details:
Experimental Method:
Resolution:
2.60 Å
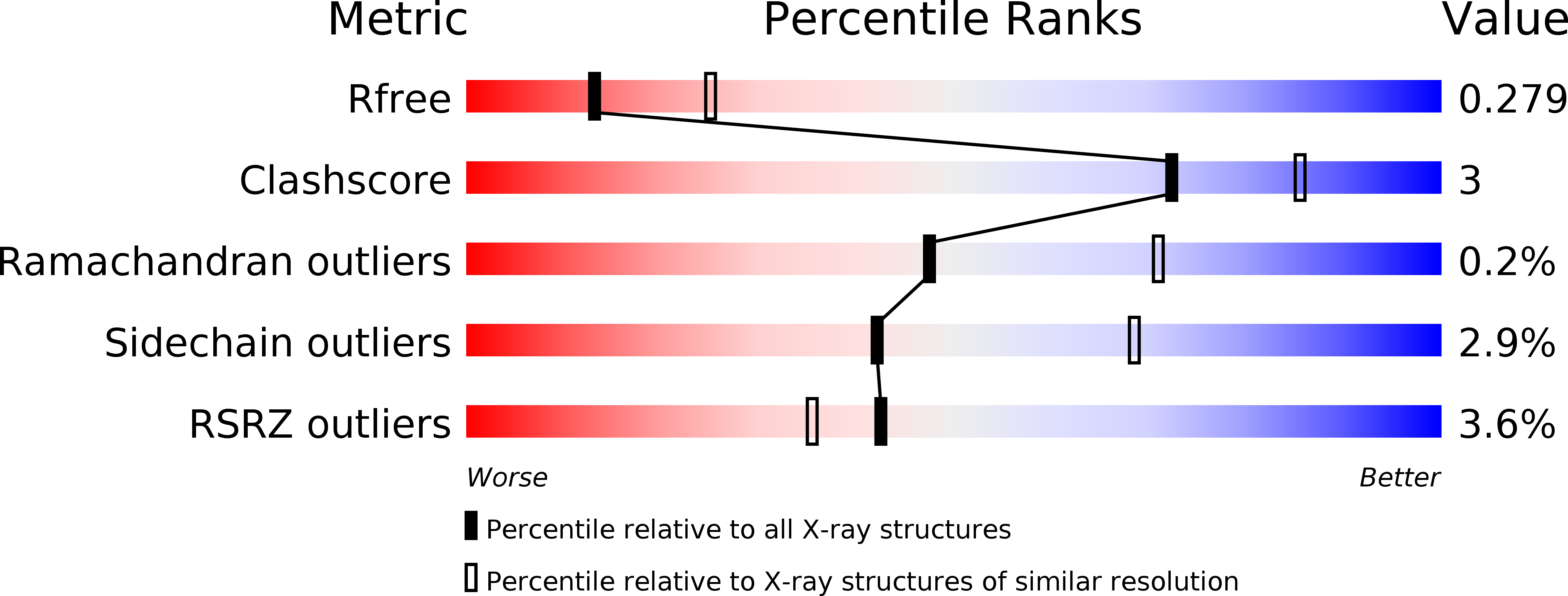
R-Value Free:
0.28
R-Value Work:
0.24
R-Value Observed:
0.24
Space Group:
P 21 21 21