Deposition Date
2012-04-19
Release Date
2012-07-04
Last Version Date
2023-12-20
Entry Detail
PDB ID:
4AQU
Keywords:
Title:
Crystal structure of I-CreI complexed with its target methylated at position plus 2 (in the b strand) in the presence of calcium
Biological Source:
Source Organism:
CHLAMYDOMONAS REINHARDTII (Taxon ID: 3055)
SYNTHETIC CONSTRUCT (Taxon ID: 32630)
SYNTHETIC CONSTRUCT (Taxon ID: 32630)
Host Organism:
Method Details:
Experimental Method:
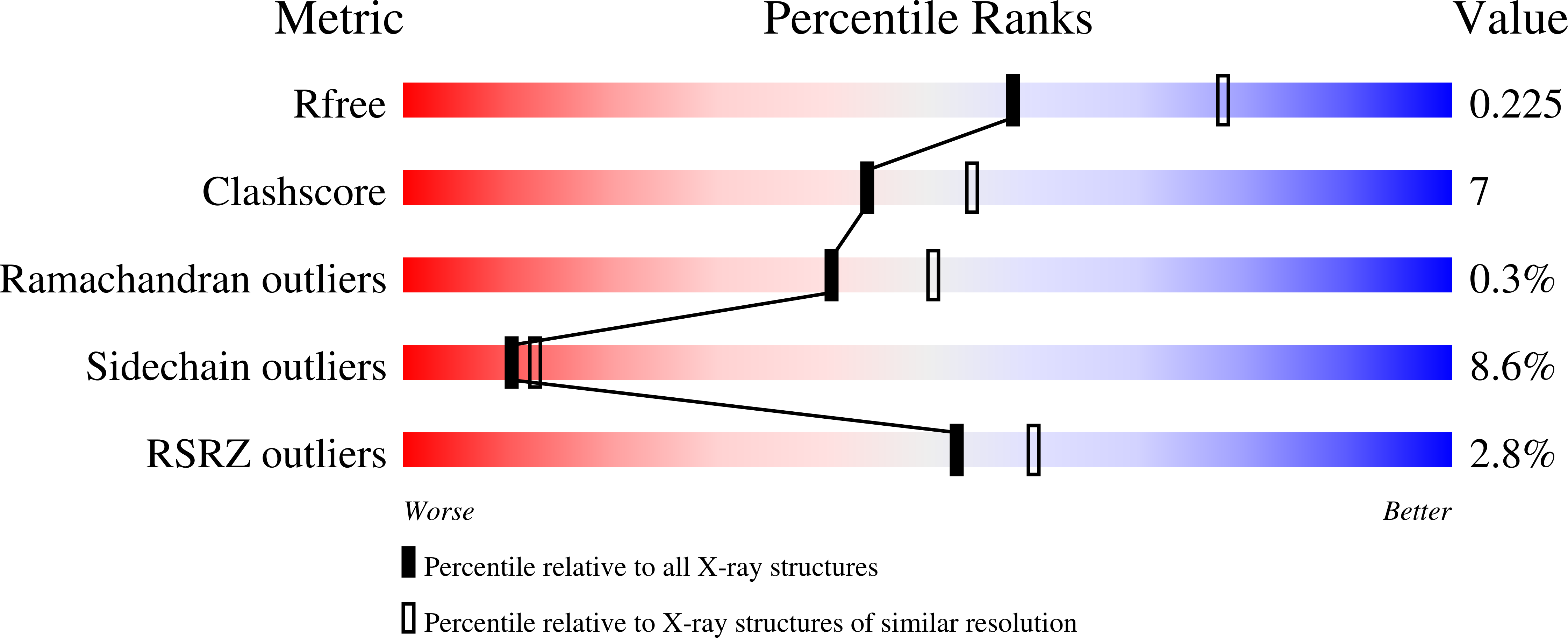
Resolution:
2.30 Å
R-Value Free:
0.24
R-Value Work:
0.19
R-Value Observed:
0.20
Space Group:
P 21 2 21