Deposition Date
2012-04-12
Release Date
2012-06-13
Last Version Date
2024-11-20
Entry Detail
PDB ID:
4AQ1
Keywords:
Title:
Structure of the SbsB S-layer protein of Geobacillus stearothermophilus PV72p2 in complex with nanobody KB6
Biological Source:
Source Organism:
GEOBACILLUS STEAROTHERMOPHILUS (Taxon ID: 1422)
LAMA GLAMA (Taxon ID: 9844)
LAMA GLAMA (Taxon ID: 9844)
Host Organism:
Method Details:
Experimental Method:
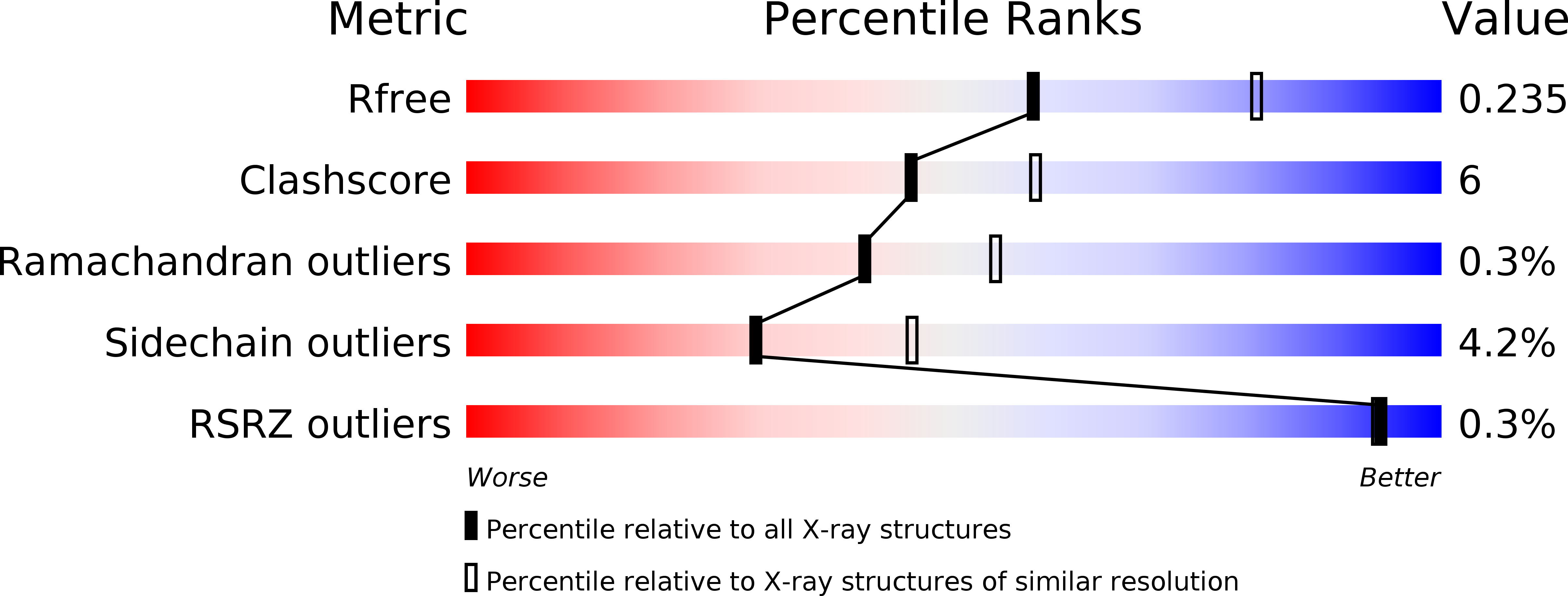
Resolution:
2.42 Å
R-Value Free:
0.23
R-Value Work:
0.18
R-Value Observed:
0.18
Space Group:
P 1